Update a wflow model: water demand#
With HydroMT, you can easily read your model and update one or several components of your model using the update function of the command line interface (CLI). Here are the steps and some examples on how to add water demand information to your wflow_sbm model.
All lines in this notebook which starts with ! are executed from the command line. Within the notebook environment the logging messages are shown after completion. You can also copy these lines and paste them in your shell to get more feedback.
You can update a wflow_sbm model to include the simulation of water demand and allocation. This can be important in basins where there is substantial human influence, and/or you might want to simulate what will happen if (more) water is consumed. We identify four different sources of water demand:
domestic
industry
livestock
irrigation
These four different items are taken into account when adding the required water demand data to your wflow model. Besides computing the water demand, we also need information on the regions where water is allocated.
Let’s open the example configuration file (wflow_update_water_demand.yml) from the model repository [examples folder] and have a look at the main setup.
[1]:
fn_config = "wflow_update_water_demand.yml"
with open(fn_config, "r") as f:
txt = f.read()
print(txt)
steps:
- setup_allocation_areas:
waterareas_fn: gadm_level2
priority_basins: True
output_name: allocation_areas_admin2
- setup_allocation_surfacewaterfrac:
gwfrac_fn: lisflood_gwfrac
gwbodies_fn: lisflood_gwbodies
ncfrac_fn: lisflood_ncfrac
# Update model with GLCNMO landuse data in order to add the paddies
- setup_lulcmaps_with_paddy:
lulc_fn: glcnmo
paddy_class: 12
soil_fn: soilgrids
wflow_thicknesslayers:
- 50
- 100
- 50
- 200
- 800
target_conductivity:
- null
- null
- 5
- null
- null
paddy_waterlevels:
"demand_paddy_h_min": 20
"demand_paddy_h_opt": 50
"demand_paddy_h_max": 80
- setup_domestic_demand:
domestic_fn: pcr_globwb
population_fn: ghs_pop_2015
domestic_fn_original_res: 0.5
- setup_other_demand:
demand_fn: pcr_globwb
variables:
- industry_gross
- industry_net
- livestock_gross
- livestock_net
resampling_method: nearest
- setup_irrigation:
irrigated_area_fn: irrigated_area
irrigation_value:
- 1
cropland_class:
- 11
- 14
- 20
- 30
paddy_class:
- 12
area_threshold: 0.6
lai_threshold: 0.2
lulcmap_name: meta_landuse
Here, you see we will be adding the required information to simulate water demand and allocation in wflow, by running three different setup functions:
setup_allocation_areas: Adds a map that defines the different regions that will be used to allocate water. By default this is a mix between administrative boundaries and catchment boundaries
setup_allocation_surfacewaterfrac: prepare the fraction of surface water used for allocation (can be reduced if groundwater or non conventional water sources are also present in the basin).
setup_lulcmaps_with_paddy (or setup_lulcmaps): update the landuse to include new parameters (crop factor and soil water pressure heads from feddes) and add the paddies (rice fields). To allow for water to pool on the surface of the rice fields, soil parameters will also be updated to include an additional thin layer with limited vertical conductivity.
setup_domestic_demand: Add domestic water demands (gross and net) from gridded data and downscaled using high resolution population map.
setup_other_demand: Adds maps to the wflow schematization that describe how much water is demanded (gross and net amounts) by different sources: domestic (dom), industry (ind), and livestock (lsk). In our case, as we downscale domestic with population, we will here add industry and livestock.
setup_irrigation: Adds information about where and when irrigation takes place for paddy and nonpaddy crops.
More information on these steps will be given below (after we have updated the model)
Update the wflow model#
First, we build a wflow_sbm model for the full Piave basin, as most water demand activities occur in the most downstream locations of the catchment. Please note that this is a basin model (until the full outlet at the ocean), as opposed to the sub-basin models (until a certain location) that are being created in the other examples.
[2]:
# NOTE: copy this line (without !) to your shell for more direct feedback
! hydromt build wflow_sbm "./wflow_piave_basin" -i wflow_build_basin.yml -d artifact_data -v
2026-01-14 11:22:02,251 - hydromt - log - INFO - HydroMT version: 1.3.0
2026-01-14 11:22:02,370 - hydromt.data_catalog.data_catalog - data_catalog - INFO - Reading data catalog artifact_data latest
2026-01-14 11:22:02,371 - hydromt.data_catalog.data_catalog - data_catalog - INFO - Parsing data catalog from /home/runner/.hydromt/artifact_data/v1.0.0/data_catalog.yml
2026-01-14 11:22:03,007 - hydromt.model.model - model - INFO - Initializing wflow_sbm model from hydromt_wflow (v1.0.1.dev0).
2026-01-14 11:22:03,007 - hydromt.data_catalog.data_catalog - data_catalog - INFO - Parsing data catalog from /home/runner/work/hydromt_wflow/hydromt_wflow/hydromt_wflow/data/parameters_data.yml
2026-01-14 11:22:03,023 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Supported Wflow.jl version v1+
2026-01-14 11:22:03,023 - hydromt.hydromt_wflow.components.config - config - INFO - Reading default config file from /home/runner/work/hydromt_wflow/hydromt_wflow/hydromt_wflow/data/wflow_sbm/wflow_sbm.toml.
2026-01-14 11:22:03,024 - hydromt - log - INFO - HydroMT version: 1.3.0
2026-01-14 11:22:03,024 - hydromt.model.model - model - INFO - build: setup_config
2026-01-14 11:22:03,024 - hydromt.model.model - model - INFO - setup_config.data={'time.starttime': datetime.datetime(2010, 1, 1, 0, 0), 'time.endtime': datetime.datetime(2010, 3, 31, 0, 0), 'time.timestepsecs': 86400, 'input.path_forcing': 'inmaps-era5-2010.nc', 'output.netcdf_grid.path': 'output.nc', 'output.netcdf_grid.compressionlevel': 1, 'output.netcdf_grid.variables.river_water__volume_flow_rate': 'river_q'}
2026-01-14 11:22:03,025 - hydromt.model.model - model - INFO - build: setup_basemaps
2026-01-14 11:22:03,025 - hydromt.model.model - model - INFO - setup_basemaps.basin_index_fn=merit_hydro_index
2026-01-14 11:22:03,025 - hydromt.model.model - model - INFO - setup_basemaps.res=0.00833
2026-01-14 11:22:03,025 - hydromt.model.model - model - INFO - setup_basemaps.upscale_method=ihu
2026-01-14 11:22:03,025 - hydromt.model.model - model - INFO - setup_basemaps.output_names={'basin__local_drain_direction': 'local_drain_direction', 'subbasin_location__count': 'subcatchment', 'land_surface__slope': 'land_slope'}
2026-01-14 11:22:03,025 - hydromt.model.model - model - INFO - setup_basemaps.hydrography_fn=merit_hydro
2026-01-14 11:22:03,025 - hydromt.model.model - model - INFO - setup_basemaps.region={'basin': [12.2051, 45.8331]}
2026-01-14 11:22:03,025 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Preparing base hydrography basemaps.
2026-01-14 11:22:03,025 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading merit_hydro RasterDataset data from /home/runner/.hydromt/artifact_data/latest/merit_hydro/{variable}.tif
2026-01-14 11:22:03,114 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading merit_hydro_index GeoDataFrame data from /home/runner/.hydromt/artifact_data/latest/merit_hydro_index.gpkg
2026-01-14 11:22:03,242 - hydromt.model.processes.basin_mask - basin_mask - INFO - basin bbox: [11.7750, 45.5042, 12.8375, 46.6900]
2026-01-14 11:22:03,308 - hydromt.hydromt_wflow.workflows.basemaps - basemaps - INFO - (Sub)basin at original resolution has 751509 cells.
2026-01-14 11:22:04,116 - hydromt.hydromt_wflow.workflows.basemaps - basemaps - INFO - Upscale flow direction data: 10x, ihu method.
2026-01-14 11:22:15,747 - hydromt.hydromt_wflow.workflows.basemaps - basemaps - INFO - Derive stream order.
2026-01-14 11:22:15,896 - hydromt.hydromt_wflow.workflows.basemaps - basemaps - INFO - Map shape: (142, 127); active cells: 7503.
2026-01-14 11:22:15,897 - hydromt.hydromt_wflow.workflows.basemaps - basemaps - INFO - Outlet coordinates (1/1): (12.73250,45.52750).
2026-01-14 11:22:15,916 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Adding basin shape to staticgeoms.
2026-01-14 11:22:15,928 - hydromt.hydromt_wflow.workflows.basemaps - basemaps - INFO - Derive elevation and slope.
2026-01-14 11:22:16,091 - hydromt.model.model - model - INFO - build: setup_rivers
2026-01-14 11:22:16,091 - hydromt.model.model - model - INFO - setup_rivers.river_geom_fn=hydro_rivers_lin
2026-01-14 11:22:16,091 - hydromt.model.model - model - INFO - setup_rivers.river_upa=30
2026-01-14 11:22:16,091 - hydromt.model.model - model - INFO - setup_rivers.rivdph_method=powlaw
2026-01-14 11:22:16,091 - hydromt.model.model - model - INFO - setup_rivers.slope_len=2000
2026-01-14 11:22:16,091 - hydromt.model.model - model - INFO - setup_rivers.min_rivlen_ratio=0.0
2026-01-14 11:22:16,091 - hydromt.model.model - model - INFO - setup_rivers.min_rivdph=1
2026-01-14 11:22:16,091 - hydromt.model.model - model - INFO - setup_rivers.min_rivwth=30
2026-01-14 11:22:16,091 - hydromt.model.model - model - INFO - setup_rivers.smooth_len=5000
2026-01-14 11:22:16,091 - hydromt.model.model - model - INFO - setup_rivers.elevtn_map=land_elevation
2026-01-14 11:22:16,092 - hydromt.model.model - model - INFO - setup_rivers.river_routing=kinematic_wave
2026-01-14 11:22:16,092 - hydromt.model.model - model - INFO - setup_rivers.connectivity=8
2026-01-14 11:22:16,092 - hydromt.model.model - model - INFO - setup_rivers.output_names={'river_location__mask': 'river_mask', 'river__length': 'river_length', 'river__width': 'river_width', 'river_bank_water__depth': 'river_depth', 'river__slope': 'river_slope', 'river_bank_water__elevation': 'river_bank_elevation'}
2026-01-14 11:22:16,092 - hydromt.model.model - model - INFO - setup_rivers.hydrography_fn=merit_hydro
2026-01-14 11:22:16,092 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Preparing river maps.
2026-01-14 11:22:16,094 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading merit_hydro RasterDataset data from /home/runner/.hydromt/artifact_data/latest/merit_hydro/{variable}.tif
2026-01-14 11:22:16,167 - hydromt.hydromt_wflow.workflows.river - river - INFO - Set river mask (min uparea: 30 km2) and prepare flow dirs.
2026-01-14 11:22:16,317 - hydromt.hydromt_wflow.workflows.river - river - INFO - Derive river length.
2026-01-14 11:22:20,442 - hydromt.hydromt_wflow.workflows.river - river - INFO - Derive river slope.
2026-01-14 11:22:20,978 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading hydro_rivers_lin GeoDataFrame data from /home/runner/.hydromt/artifact_data/latest/rivers_lin2019_v1.gpkg
2026-01-14 11:22:21,133 - hydromt.hydromt_wflow.workflows.river - river - INFO - Derive ['rivwth', 'qbankfull'] from shapefile.
2026-01-14 11:22:21,202 - hydromt.hydromt_wflow.workflows.river - river - INFO - Valid for 466/830 river cells (max dist: 393 m).
2026-01-14 11:22:23,782 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Update wflow config model.river_routing="kinematic_wave"
2026-01-14 11:22:23,783 - hydromt.model.model - model - INFO - build: setup_river_roughness
2026-01-14 11:22:23,783 - hydromt.model.model - model - INFO - setup_river_roughness.rivman_mapping_fn=None
2026-01-14 11:22:23,783 - hydromt.model.model - model - INFO - setup_river_roughness.strord_name=meta_streamorder
2026-01-14 11:22:23,783 - hydromt.model.model - model - INFO - setup_river_roughness.output_name=river_manning_n
2026-01-14 11:22:23,783 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing river Manning roughness.
2026-01-14 11:22:23,783 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading roughness_river_mapping_default DataFrame data from /home/runner/work/hydromt_wflow/hydromt_wflow/hydromt_wflow/data/wflow_sbm/river_manning_mapping.csv
2026-01-14 11:22:23,789 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving river_manning_n using average resampling (nodata=-999.0).
2026-01-14 11:22:23,796 - hydromt.model.model - model - INFO - build: setup_reservoirs_simple_control
2026-01-14 11:22:23,796 - hydromt.model.model - model - INFO - setup_reservoirs_simple_control.timeseries_fn=gww
2026-01-14 11:22:23,796 - hydromt.model.model - model - INFO - setup_reservoirs_simple_control.overwrite_existing=False
2026-01-14 11:22:23,796 - hydromt.model.model - model - INFO - setup_reservoirs_simple_control.duplicate_id=error
2026-01-14 11:22:23,796 - hydromt.model.model - model - INFO - setup_reservoirs_simple_control.min_area=1.0
2026-01-14 11:22:23,796 - hydromt.model.model - model - INFO - setup_reservoirs_simple_control.output_names={'reservoir_area__count': 'reservoir_area_id', 'reservoir_location__count': 'reservoir_outlet_id', 'reservoir_surface__area': 'reservoir_area', 'reservoir_water_surface__initial_elevation': 'reservoir_initial_depth', 'reservoir_water__rating_curve_type_count': 'reservoir_rating_curve', 'reservoir_water__storage_curve_type_count': 'reservoir_storage_curve', 'reservoir_water__max_volume': 'reservoir_max_volume', 'reservoir_water__target_min_volume_fraction': 'reservoir_target_min_fraction', 'reservoir_water__target_full_volume_fraction': 'reservoir_target_full_fraction', 'reservoir_water_demand__required_downstream_volume_flow_rate': 'reservoir_demand', 'reservoir_water_release_below_spillway__max_volume_flow_rate': 'reservoir_max_release'}
2026-01-14 11:22:23,796 - hydromt.model.model - model - INFO - setup_reservoirs_simple_control.geom_name=meta_reservoirs_simple_control
2026-01-14 11:22:23,796 - hydromt.model.model - model - INFO - setup_reservoirs_simple_control.reservoirs_fn=hydro_reservoirs
2026-01-14 11:22:23,797 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing reservoir with simple control maps.
2026-01-14 11:22:23,797 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading hydro_reservoirs GeoDataFrame data from /home/runner/.hydromt/artifact_data/latest/hydro_reservoirs.gpkg
2026-01-14 11:22:23,814 - hydromt.hydromt_wflow.workflows.reservoirs - reservoirs - INFO - 0 reservoir(s) of sufficient size found within region.
2026-01-14 11:22:23,814 - hydromt.model.model - model - INFO - build: setup_reservoirs_no_control
2026-01-14 11:22:23,814 - hydromt.model.model - model - INFO - setup_reservoirs_no_control.rating_curve_fns=None
2026-01-14 11:22:23,814 - hydromt.model.model - model - INFO - setup_reservoirs_no_control.overwrite_existing=False
2026-01-14 11:22:23,814 - hydromt.model.model - model - INFO - setup_reservoirs_no_control.duplicate_id=error
2026-01-14 11:22:23,814 - hydromt.model.model - model - INFO - setup_reservoirs_no_control.min_area=10.0
2026-01-14 11:22:23,814 - hydromt.model.model - model - INFO - setup_reservoirs_no_control.output_names={'reservoir_area__count': 'reservoir_area_id', 'reservoir_location__count': 'reservoir_outlet_id', 'reservoir_surface__area': 'reservoir_area', 'reservoir_water_surface__initial_elevation': 'reservoir_initial_depth', 'reservoir_water_flow_threshold_level__elevation': 'reservoir_outflow_threshold', 'reservoir_water__rating_curve_coefficient': 'reservoir_b', 'reservoir_water__rating_curve_exponent': 'reservoir_e', 'reservoir_water__rating_curve_type_count': 'reservoir_rating_curve', 'reservoir_water__storage_curve_type_count': 'reservoir_storage_curve', 'reservoir_lower_location__count': 'reservoir_lower_id'}
2026-01-14 11:22:23,814 - hydromt.model.model - model - INFO - setup_reservoirs_no_control.geom_name=meta_reservoirs_no_control
2026-01-14 11:22:23,814 - hydromt.model.model - model - INFO - setup_reservoirs_no_control.reservoirs_fn=hydro_lakes
2026-01-14 11:22:23,814 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing reservoir maps.
2026-01-14 11:22:23,815 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading hydro_lakes GeoDataFrame data from /home/runner/.hydromt/artifact_data/latest/hydro_lakes.gpkg
2026-01-14 11:22:23,824 - hydromt.hydromt_wflow.workflows.reservoirs - reservoirs - INFO - 0 reservoir(s) of sufficient size found within region.
2026-01-14 11:22:23,824 - hydromt.model.model - model - INFO - build: setup_glaciers
2026-01-14 11:22:23,825 - hydromt.model.model - model - INFO - setup_glaciers.min_area=0.0
2026-01-14 11:22:23,825 - hydromt.model.model - model - INFO - setup_glaciers.output_names={'glacier_surface__area_fraction': 'glacier_fraction', 'glacier_ice__initial_leq_depth': 'glacier_initial_leq_depth'}
2026-01-14 11:22:23,825 - hydromt.model.model - model - INFO - setup_glaciers.geom_name=glaciers
2026-01-14 11:22:23,825 - hydromt.model.model - model - INFO - setup_glaciers.glaciers_fn=rgi
2026-01-14 11:22:23,825 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing glacier maps.
2026-01-14 11:22:23,825 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading rgi GeoDataFrame data from /home/runner/.hydromt/artifact_data/latest/rgi.gpkg
2026-01-14 11:22:23,835 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - 38 glaciers of sufficient size found within region.
2026-01-14 11:22:24,659 - hydromt.model.model - model - INFO - build: setup_lulcmaps
2026-01-14 11:22:24,660 - hydromt.model.model - model - INFO - setup_lulcmaps.lulc_mapping_fn=globcover_mapping_default
2026-01-14 11:22:24,660 - hydromt.model.model - model - INFO - setup_lulcmaps.lulc_vars=['landuse', 'vegetation_kext', 'land_manning_n', 'soil_compacted_fraction', 'vegetation_root_depth', 'vegetation_leaf_storage', 'vegetation_wood_storage', 'land_water_fraction', 'vegetation_crop_factor', 'vegetation_feddes_alpha_h1', 'vegetation_feddes_h1', 'vegetation_feddes_h2', 'vegetation_feddes_h3_high', 'vegetation_feddes_h3_low', 'vegetation_feddes_h4']
2026-01-14 11:22:24,660 - hydromt.model.model - model - INFO - setup_lulcmaps.output_names_suffix=None
2026-01-14 11:22:24,660 - hydromt.model.model - model - INFO - setup_lulcmaps.lulc_fn=globcover_2009
2026-01-14 11:22:24,660 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Preparing LULC parameter maps.
2026-01-14 11:22:24,661 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading globcover_2009 RasterDataset data from /home/runner/.hydromt/artifact_data/latest/globcover.tif
2026-01-14 11:22:24,671 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading globcover_mapping_default DataFrame data from /home/runner/work/hydromt_wflow/hydromt_wflow/hydromt_wflow/data/lulc/v1.0/globcover_mapping.csv
2026-01-14 11:22:24,678 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving landuse using mode resampling (nodata=230).
2026-01-14 11:22:24,761 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_kext using average resampling (nodata=-999.0).
2026-01-14 11:22:24,784 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving land_manning_n using average resampling (nodata=-999.0).
2026-01-14 11:22:24,806 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving soil_compacted_fraction using average resampling (nodata=-999.0).
2026-01-14 11:22:24,829 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_root_depth using average resampling (nodata=-999.0).
2026-01-14 11:22:24,851 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_leaf_storage using average resampling (nodata=-999.0).
2026-01-14 11:22:24,875 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_wood_storage using average resampling (nodata=-999.0).
2026-01-14 11:22:24,897 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving land_water_fraction using average resampling (nodata=-999.0).
2026-01-14 11:22:24,920 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_crop_factor using average resampling (nodata=-999.0).
2026-01-14 11:22:24,944 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_alpha_h1 using mode resampling (nodata=-999).
2026-01-14 11:22:25,025 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_h1 using average resampling (nodata=-999).
2026-01-14 11:22:25,051 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_h2 using average resampling (nodata=-999).
2026-01-14 11:22:25,078 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_h3_high using average resampling (nodata=-999).
2026-01-14 11:22:25,105 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_h3_low using average resampling (nodata=-999).
2026-01-14 11:22:25,132 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_h4 using average resampling (nodata=-999).
2026-01-14 11:22:25,186 - hydromt.model.model - model - INFO - build: setup_laimaps
2026-01-14 11:22:25,186 - hydromt.model.model - model - INFO - setup_laimaps.lulc_fn=None
2026-01-14 11:22:25,186 - hydromt.model.model - model - INFO - setup_laimaps.lulc_sampling_method=any
2026-01-14 11:22:25,186 - hydromt.model.model - model - INFO - setup_laimaps.lulc_zero_classes=[]
2026-01-14 11:22:25,186 - hydromt.model.model - model - INFO - setup_laimaps.buffer=2
2026-01-14 11:22:25,186 - hydromt.model.model - model - INFO - setup_laimaps.output_name=vegetation_leaf_area_index
2026-01-14 11:22:25,186 - hydromt.model.model - model - INFO - setup_laimaps.lai_fn=modis_lai
2026-01-14 11:22:25,186 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing LAI maps.
2026-01-14 11:22:25,188 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading modis_lai RasterDataset data from /home/runner/.hydromt/artifact_data/latest/modis_lai.nc
2026-01-14 11:22:25,749 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving LAI using average resampling (nodata=nan).
2026-01-14 11:22:25,823 - hydromt.model.model - model - INFO - build: setup_soilmaps
2026-01-14 11:22:25,824 - hydromt.model.model - model - INFO - setup_soilmaps.soil_fn=soilgrids
2026-01-14 11:22:25,824 - hydromt.model.model - model - INFO - setup_soilmaps.ptf_ksatver=brakensiek
2026-01-14 11:22:25,824 - hydromt.model.model - model - INFO - setup_soilmaps.wflow_thicknesslayers=[100, 300, 800]
2026-01-14 11:22:25,824 - hydromt.model.model - model - INFO - setup_soilmaps.output_names={'soil_water__saturated_volume_fraction': 'soil_theta_s', 'soil_water__residual_volume_fraction': 'soil_theta_r', 'soil_surface_water__vertical_saturated_hydraulic_conductivity': 'soil_ksat_vertical', 'soil__thickness': 'soil_thickness', 'soil_water__vertical_saturated_hydraulic_conductivity_scale_parameter': 'soil_f', 'soil_layer_water__brooks_corey_exponent': 'soil_brooks_corey_c'}
2026-01-14 11:22:25,824 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing soil parameter maps.
2026-01-14 11:22:25,826 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading soilgrids RasterDataset data from /home/runner/.hydromt/artifact_data/latest/soilgrids/{variable}.tif
2026-01-14 11:22:26,342 - hydromt.hydromt_wflow.workflows.soilgrids - soilgrids - INFO - calculate and resample theta_s
2026-01-14 11:22:26,544 - hydromt.hydromt_wflow.workflows.soilgrids - soilgrids - INFO - calculate and resample theta_r
2026-01-14 11:22:26,792 - hydromt.hydromt_wflow.workflows.soilgrids - soilgrids - INFO - calculate and resample ksat_vertical
2026-01-14 11:22:26,851 - hydromt.hydromt_wflow.workflows.soilgrids - soilgrids - INFO - calculate and resample pore size distribution index
2026-01-14 11:22:27,333 - hydromt.hydromt_wflow.workflows.soilgrids - soilgrids - INFO - fit z - log(ksat_vertical ) with numpy linalg regression (y = b*x) -> M_
2026-01-14 11:22:27,851 - hydromt.hydromt_wflow.workflows.soilgrids - soilgrids - INFO - fit zi - Ksat with curve_fit (scipy.optimize) -> M
2026-01-14 11:22:32,370 - hydromt.model.model - model - INFO - build: setup_outlets
2026-01-14 11:22:32,370 - hydromt.model.model - model - INFO - setup_outlets.river_only=True
2026-01-14 11:22:32,371 - hydromt.model.model - model - INFO - setup_outlets.toml_output=csv
2026-01-14 11:22:32,371 - hydromt.model.model - model - INFO - setup_outlets.gauge_toml_header=['river_q']
2026-01-14 11:22:32,371 - hydromt.model.model - model - INFO - setup_outlets.gauge_toml_param=['river_water__volume_flow_rate']
2026-01-14 11:22:32,371 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Gauges locations set based on river outlets.
2026-01-14 11:22:32,383 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Gauges map based on catchment river outlets added.
2026-01-14 11:22:32,383 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Adding ['river_water__volume_flow_rate'] to csv section of toml.
2026-01-14 11:22:32,383 - hydromt.model.model - model - INFO - build: setup_gauges
2026-01-14 11:22:32,383 - hydromt.model.model - model - INFO - setup_gauges.index_col=None
2026-01-14 11:22:32,383 - hydromt.model.model - model - INFO - setup_gauges.snap_to_river=True
2026-01-14 11:22:32,383 - hydromt.model.model - model - INFO - setup_gauges.mask=None
2026-01-14 11:22:32,383 - hydromt.model.model - model - INFO - setup_gauges.snap_uparea=False
2026-01-14 11:22:32,383 - hydromt.model.model - model - INFO - setup_gauges.max_dist=10000.0
2026-01-14 11:22:32,383 - hydromt.model.model - model - INFO - setup_gauges.wdw=3
2026-01-14 11:22:32,383 - hydromt.model.model - model - INFO - setup_gauges.rel_error=0.05
2026-01-14 11:22:32,383 - hydromt.model.model - model - INFO - setup_gauges.abs_error=50.0
2026-01-14 11:22:32,384 - hydromt.model.model - model - INFO - setup_gauges.fillna=False
2026-01-14 11:22:32,384 - hydromt.model.model - model - INFO - setup_gauges.derive_subcatch=False
2026-01-14 11:22:32,384 - hydromt.model.model - model - INFO - setup_gauges.basename=None
2026-01-14 11:22:32,384 - hydromt.model.model - model - INFO - setup_gauges.toml_output=csv
2026-01-14 11:22:32,384 - hydromt.model.model - model - INFO - setup_gauges.gauge_toml_header=['river_q', 'precip']
2026-01-14 11:22:32,384 - hydromt.model.model - model - INFO - setup_gauges.gauge_toml_param=['river_water__volume_flow_rate', 'atmosphere_water__precipitation_volume_flux']
2026-01-14 11:22:32,384 - hydromt.model.model - model - INFO - setup_gauges.gauges_fn=grdc
2026-01-14 11:22:32,384 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading grdc GeoDataFrame data from /home/runner/.hydromt/artifact_data/latest/grdc.csv
2026-01-14 11:22:32,390 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - 3 grdc gauge locations found within domain
2026-01-14 11:22:32,940 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Adding ['river_water__volume_flow_rate', 'atmosphere_water__precipitation_volume_flux'] to csv section of toml.
2026-01-14 11:22:32,940 - hydromt.model.model - model - INFO - build: setup_precip_forcing
2026-01-14 11:22:32,940 - hydromt.model.model - model - INFO - setup_precip_forcing.precip_clim_fn=None
2026-01-14 11:22:32,940 - hydromt.model.model - model - INFO - setup_precip_forcing.chunksize=None
2026-01-14 11:22:32,940 - hydromt.model.model - model - INFO - setup_precip_forcing.precip_fn=era5
2026-01-14 11:22:32,942 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading era5 RasterDataset data from /home/runner/.hydromt/artifact_data/latest/era5.nc
2026-01-14 11:22:33,018 - hydromt.model.model - model - INFO - build: setup_temp_pet_forcing
2026-01-14 11:22:33,019 - hydromt.model.model - model - INFO - setup_temp_pet_forcing.pet_method=debruin
2026-01-14 11:22:33,019 - hydromt.model.model - model - INFO - setup_temp_pet_forcing.press_correction=True
2026-01-14 11:22:33,019 - hydromt.model.model - model - INFO - setup_temp_pet_forcing.temp_correction=True
2026-01-14 11:22:33,019 - hydromt.model.model - model - INFO - setup_temp_pet_forcing.wind_correction=True
2026-01-14 11:22:33,019 - hydromt.model.model - model - INFO - setup_temp_pet_forcing.wind_altitude=10
2026-01-14 11:22:33,019 - hydromt.model.model - model - INFO - setup_temp_pet_forcing.reproj_method=nearest_index
2026-01-14 11:22:33,019 - hydromt.model.model - model - INFO - setup_temp_pet_forcing.fillna_method=None
2026-01-14 11:22:33,019 - hydromt.model.model - model - INFO - setup_temp_pet_forcing.dem_forcing_fn=era5_orography
2026-01-14 11:22:33,019 - hydromt.model.model - model - INFO - setup_temp_pet_forcing.skip_pet=False
2026-01-14 11:22:33,019 - hydromt.model.model - model - INFO - setup_temp_pet_forcing.chunksize=None
2026-01-14 11:22:33,019 - hydromt.model.model - model - INFO - setup_temp_pet_forcing.temp_pet_fn=era5
2026-01-14 11:22:33,021 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading era5 RasterDataset data from /home/runner/.hydromt/artifact_data/latest/era5.nc
2026-01-14 11:22:33,065 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading era5_orography RasterDataset data from /home/runner/.hydromt/artifact_data/latest/era5_orography.nc
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - build: setup_constant_pars
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - setup_constant_pars.subsurface_water__horizontal_to_vertical_saturated_hydraulic_conductivity_ratio=100
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - setup_constant_pars.snowpack__degree_day_coefficient=3.75653
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - setup_constant_pars.soil_surface_water__infiltration_reduction_parameter=0.038
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - setup_constant_pars.vegetation_canopy_water__mean_evaporation_to_mean_precipitation_ratio=0.11
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - setup_constant_pars.compacted_soil_surface_water__infiltration_capacity=10
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - setup_constant_pars.soil_water_saturated_zone_bottom__max_leakage_volume_flux=0
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - setup_constant_pars.soil_wet_root__sigmoid_function_shape_parameter=-500
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - setup_constant_pars.atmosphere_air__snowfall_temperature_threshold=0
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - setup_constant_pars.atmosphere_air__snowfall_temperature_interval=1
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - setup_constant_pars.snowpack__melting_temperature_threshold=0
2026-01-14 11:22:33,300 - hydromt.model.model - model - INFO - setup_constant_pars.snowpack__liquid_water_holding_capacity=0.1
2026-01-14 11:22:33,301 - hydromt.model.model - model - INFO - setup_constant_pars.glacier_ice__degree_day_coefficient=3.0
2026-01-14 11:22:33,301 - hydromt.model.model - model - INFO - setup_constant_pars.glacier_firn_accumulation__snowpack_dry_snow_leq_depth_fraction=0.001
2026-01-14 11:22:33,301 - hydromt.model.model - model - INFO - setup_constant_pars.glacier_ice__melting_temperature_threshold=0.0
2026-01-14 11:22:33,301 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Write model data to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin
2026-01-14 11:22:33,311 - hydromt.model.components.grid - grid - INFO - wflow_sbm.staticmaps: Writing grid data to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/staticmaps.nc.
2026-01-14 11:22:33,414 - hydromt.model.components.spatial - spatial - INFO - wflow_sbm.staticmaps: Writing region to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/staticgeoms/region.geojson.
2026-01-14 11:22:33,441 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/staticgeoms/meta_basins_highres.geojson.
2026-01-14 11:22:33,455 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/staticgeoms/basins.geojson.
2026-01-14 11:22:33,458 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/staticgeoms/rivers.geojson.
2026-01-14 11:22:33,462 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/staticgeoms/glaciers.geojson.
2026-01-14 11:22:33,483 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/staticgeoms/outlets.geojson.
2026-01-14 11:22:33,485 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/staticgeoms/gauges_grdc.geojson.
2026-01-14 11:22:33,488 - hydromt.hydromt_wflow.components.forcing - forcing - INFO - Write forcing file
2026-01-14 11:22:33,488 - hydromt.hydromt_wflow.components.forcing - forcing - WARNING - Start time 2010-01-01T00:00:00.000000 does not match the beginning of the data. Changing to start of the data: 2010-02-02T00:00:00.000000000.
2026-01-14 11:22:33,488 - hydromt.hydromt_wflow.components.forcing - forcing - WARNING - End time 2010-03-31T00:00:00.000000 does not match the end of the data. Changing to end of the data: 2010-02-15T00:00:00.000000000.
2026-01-14 11:22:33,490 - hydromt.hydromt_wflow.components.forcing - forcing - INFO - Writing file /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/inmaps-era5-2010.nc
[########################################] | 100% Completed | 101.32 ms
2026-01-14 11:22:33,613 - hydromt.model.components.tables - tables - INFO - wflow_sbm.tables: No tables found, skip writing.
2026-01-14 11:22:33,613 - hydromt.model.components.grid - grid - INFO - wflow_sbm.states: No grid data found, skip writing.
2026-01-14 11:22:33,614 - hydromt.hydromt_wflow.components.config - config - INFO - Writing model config to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/wflow_sbm.toml.
Next, we will update the model using the same configuration file that was shown earlier:
[3]:
! hydromt update wflow_sbm "./wflow_piave_basin" -o "./wflow_piave_water_demand" -i wflow_update_water_demand.yml -d artifact_data -d ./data/demand/data_catalog.yml -v
2026-01-14 11:22:36,408 - hydromt - log - INFO - HydroMT version: 1.3.0
2026-01-14 11:22:36,523 - hydromt.data_catalog.data_catalog - data_catalog - INFO - Reading data catalog artifact_data latest
2026-01-14 11:22:36,523 - hydromt.data_catalog.data_catalog - data_catalog - INFO - Parsing data catalog from /home/runner/.hydromt/artifact_data/v1.0.0/data_catalog.yml
2026-01-14 11:22:37,160 - hydromt.data_catalog.data_catalog - data_catalog - INFO - Parsing data catalog from ./data/demand/data_catalog.yml
2026-01-14 11:22:37,168 - hydromt.model.model - model - INFO - Initializing wflow_sbm model from hydromt_wflow (v1.0.1.dev0).
2026-01-14 11:22:37,168 - hydromt.data_catalog.data_catalog - data_catalog - INFO - Parsing data catalog from /home/runner/work/hydromt_wflow/hydromt_wflow/hydromt_wflow/data/parameters_data.yml
2026-01-14 11:22:37,184 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Supported Wflow.jl version v1+
2026-01-14 11:22:37,184 - hydromt.hydromt_wflow.components.config - config - INFO - Reading model config file from /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/wflow_sbm.toml.
2026-01-14 11:22:37,185 - hydromt - log - INFO - HydroMT version: 1.3.0
2026-01-14 11:22:37,185 - hydromt.hydromt_wflow.components.config - config - INFO - Reading model config file from /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_basin/wflow_sbm.toml.
2026-01-14 11:22:37,913 - hydromt.hydromt_wflow.components.states - states - WARNING - CRS not found in states data, setting to model CRS.
2026-01-14 11:22:37,917 - hydromt.hydromt_wflow.components.tables - tables - INFO - Reading model table files.
2026-01-14 11:22:37,917 - hydromt - log - INFO - HydroMT version: 1.3.0
2026-01-14 11:22:37,919 - hydromt.model.model - model - INFO - update: setup_allocation_areas
2026-01-14 11:22:37,919 - hydromt.model.model - model - INFO - setup_allocation_areas.priority_basins=True
2026-01-14 11:22:37,919 - hydromt.model.model - model - INFO - setup_allocation_areas.minimum_area=50.0
2026-01-14 11:22:37,919 - hydromt.model.model - model - INFO - setup_allocation_areas.output_name=allocation_areas_admin2
2026-01-14 11:22:37,919 - hydromt.model.model - model - INFO - setup_allocation_areas.waterareas_fn=gadm_level2
2026-01-14 11:22:37,919 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing water demand allocation map.
2026-01-14 11:22:37,921 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading gadm_level2 GeoDataFrame data from /home/runner/.hydromt/artifact_data/latest/gadm_level2.gpkg
2026-01-14 11:22:38,046 - hydromt.model.model - model - INFO - update: setup_allocation_surfacewaterfrac
2026-01-14 11:22:38,046 - hydromt.model.model - model - INFO - setup_allocation_surfacewaterfrac.waterareas_fn=None
2026-01-14 11:22:38,046 - hydromt.model.model - model - INFO - setup_allocation_surfacewaterfrac.gwbodies_fn=lisflood_gwbodies
2026-01-14 11:22:38,046 - hydromt.model.model - model - INFO - setup_allocation_surfacewaterfrac.ncfrac_fn=lisflood_ncfrac
2026-01-14 11:22:38,046 - hydromt.model.model - model - INFO - setup_allocation_surfacewaterfrac.interpolate_nodata=False
2026-01-14 11:22:38,046 - hydromt.model.model - model - INFO - setup_allocation_surfacewaterfrac.mask_and_scale_gwfrac=True
2026-01-14 11:22:38,047 - hydromt.model.model - model - INFO - setup_allocation_surfacewaterfrac.output_name=demand_surface_water_ratio
2026-01-14 11:22:38,047 - hydromt.model.model - model - INFO - setup_allocation_surfacewaterfrac.gwfrac_fn=lisflood_gwfrac
2026-01-14 11:22:38,047 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing surface water fraction map.
2026-01-14 11:22:38,048 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading lisflood_gwfrac RasterDataset data from /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/data/demand/lisflood_gwfrac.tif
2026-01-14 11:22:38,075 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading lisflood_gwbodies RasterDataset data from /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/data/demand/lisflood_gwbodies.tif
2026-01-14 11:22:38,086 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading lisflood_ncfrac RasterDataset data from /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/data/demand/lisflood_ncfrac.tif
2026-01-14 11:22:38,095 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Using wflow model allocation areas.
2026-01-14 11:22:38,269 - hydromt.model.model - model - INFO - update: setup_lulcmaps_with_paddy
2026-01-14 11:22:38,269 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.output_paddy_class=None
2026-01-14 11:22:38,269 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.lulc_mapping_fn=None
2026-01-14 11:22:38,269 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.paddy_fn=None
2026-01-14 11:22:38,269 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.paddy_mapping_fn=None
2026-01-14 11:22:38,269 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.soil_fn=soilgrids
2026-01-14 11:22:38,269 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.wflow_thicknesslayers=[50, 100, 50, 200, 800]
2026-01-14 11:22:38,269 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.target_conductivity=[None, None, 5, None, None]
2026-01-14 11:22:38,269 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.lulc_vars=['landuse', 'vegetation_kext', 'land_manning_n', 'soil_compacted_fraction', 'vegetation_root_depth', 'vegetation_leaf_storage', 'vegetation_wood_storage', 'land_water_fraction', 'vegetation_crop_factor', 'vegetation_feddes_alpha_h1', 'vegetation_feddes_h1', 'vegetation_feddes_h2', 'vegetation_feddes_h3_high', 'vegetation_feddes_h3_low', 'vegetation_feddes_h4']
2026-01-14 11:22:38,269 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.paddy_waterlevels={'demand_paddy_h_min': 20, 'demand_paddy_h_opt': 50, 'demand_paddy_h_max': 80}
2026-01-14 11:22:38,269 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.save_high_resolution_lulc=False
2026-01-14 11:22:38,270 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.output_names_suffix=None
2026-01-14 11:22:38,270 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.lulc_fn=glcnmo
2026-01-14 11:22:38,270 - hydromt.model.model - model - INFO - setup_lulcmaps_with_paddy.paddy_class=12
2026-01-14 11:22:38,270 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing LULC parameter maps including paddies.
2026-01-14 11:22:38,271 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading glcnmo RasterDataset data from /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/data/demand/glcnmo.tif
2026-01-14 11:22:38,281 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading glcnmo_mapping_default DataFrame data from /home/runner/work/hydromt_wflow/hydromt_wflow/hydromt_wflow/data/lulc/v1.0/glcnmo_mapping.csv
2026-01-14 11:22:38,289 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving landuse using mode resampling (nodata=0).
2026-01-14 11:22:38,367 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_kext using average resampling (nodata=-999.0).
2026-01-14 11:22:38,383 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving land_manning_n using average resampling (nodata=-999.0).
2026-01-14 11:22:38,399 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving soil_compacted_fraction using average resampling (nodata=-999.0).
2026-01-14 11:22:38,414 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_root_depth using average resampling (nodata=-999.0).
2026-01-14 11:22:38,432 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_leaf_storage using average resampling (nodata=-999.0).
2026-01-14 11:22:38,448 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_wood_storage using average resampling (nodata=-999.0).
2026-01-14 11:22:38,463 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving land_water_fraction using average resampling (nodata=-999.0).
2026-01-14 11:22:38,479 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_crop_factor using average resampling (nodata=-999.0).
2026-01-14 11:22:38,495 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_alpha_h1 using mode resampling (nodata=-999).
2026-01-14 11:22:38,565 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_h1 using average resampling (nodata=-999).
2026-01-14 11:22:38,582 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_h2 using average resampling (nodata=-999).
2026-01-14 11:22:38,600 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_h3_high using average resampling (nodata=-999).
2026-01-14 11:22:38,618 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_h3_low using average resampling (nodata=-999).
2026-01-14 11:22:38,635 - hydromt.hydromt_wflow.workflows.landuse - landuse - INFO - Deriving vegetation_feddes_h4 using average resampling (nodata=-999).
2026-01-14 11:22:38,654 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: meta_landuse
2026-01-14 11:22:38,656 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: vegetation_kext
2026-01-14 11:22:38,659 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: land_manning_n
2026-01-14 11:22:38,661 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: soil_compacted_fraction
2026-01-14 11:22:38,663 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: vegetation_root_depth
2026-01-14 11:22:38,665 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: vegetation_leaf_storage
2026-01-14 11:22:38,668 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: vegetation_wood_storage
2026-01-14 11:22:38,670 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: land_water_fraction
2026-01-14 11:22:38,672 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: vegetation_crop_factor
2026-01-14 11:22:38,674 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: vegetation_feddes_alpha_h1
2026-01-14 11:22:38,676 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: vegetation_feddes_h1
2026-01-14 11:22:38,679 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: vegetation_feddes_h2
2026-01-14 11:22:38,681 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: vegetation_feddes_h3_high
2026-01-14 11:22:38,683 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: vegetation_feddes_h3_low
2026-01-14 11:22:38,685 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - INFO - Replacing grid map: vegetation_feddes_h4
2026-01-14 11:22:38,688 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Different thicknesslayers requested, updating `soil_brooks_corey_c` parameter
2026-01-14 11:22:38,690 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading soilgrids RasterDataset data from /home/runner/.hydromt/artifact_data/latest/soilgrids/{variable}.tif
2026-01-14 11:22:39,063 - hydromt.hydromt_wflow.workflows.soilgrids - soilgrids - INFO - Adding soil_ksat_vertical_factor map
2026-01-14 11:22:39,206 - hydromt.hydromt_wflow.workflows.soilgrids - soilgrids - INFO - calculate and resample theta_s
2026-01-14 11:22:39,208 - hydromt.hydromt_wflow.workflows.soilgrids - soilgrids - INFO - calculate and resample pore size distribution index
2026-01-14 11:22:39,955 - hydromt.hydromt_wflow.components.staticmaps - staticmaps - WARNING - Replacing 'layer' coordinate, dropping variables (['soil_brooks_corey_c']) associated with old coordinate
2026-01-14 11:22:39,962 - hydromt.model.model - model - INFO - update: setup_domestic_demand
2026-01-14 11:22:39,962 - hydromt.model.model - model - INFO - setup_domestic_demand.population_fn=ghs_pop_2015
2026-01-14 11:22:39,962 - hydromt.model.model - model - INFO - setup_domestic_demand.domestic_fn_original_res=0.5
2026-01-14 11:22:39,962 - hydromt.model.model - model - INFO - setup_domestic_demand.output_names={'domestic__gross_water_demand_volume_flux': 'demand_domestic_gross', 'domestic__net_water_demand_volume_flux': 'demand_domestic_net'}
2026-01-14 11:22:39,962 - hydromt.model.model - model - INFO - setup_domestic_demand.domestic_fn=pcr_globwb
2026-01-14 11:22:39,963 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing domestic demand maps.
2026-01-14 11:22:39,967 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading pcr_globwb RasterDataset data from /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/data/demand/pcr_globwb.nc
2026-01-14 11:22:39,991 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading pcr_globwb RasterDataset data from /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/data/demand/pcr_globwb.nc
2026-01-14 11:22:40,013 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading ghs_pop_2015 RasterDataset data from /home/runner/.hydromt/artifact_data/latest/ghs_pop_2015.tif
2026-01-14 11:22:40,790 - hydromt.model.model - model - INFO - update: setup_other_demand
2026-01-14 11:22:40,790 - hydromt.model.model - model - INFO - setup_other_demand.variables=['industry_gross', 'industry_net', 'livestock_gross', 'livestock_net']
2026-01-14 11:22:40,790 - hydromt.model.model - model - INFO - setup_other_demand.resampling_method=nearest
2026-01-14 11:22:40,790 - hydromt.model.model - model - INFO - setup_other_demand.output_names={'industry__gross_water_demand_volume_flux': 'demand_industry_gross', 'industry__net_water_demand_volume_flux': 'demand_industry_net', 'livestock__gross_water_demand_volume_flux': 'demand_livestock_gross', 'livestock__net_water_demand_volume_flux': 'demand_livestock_net'}
2026-01-14 11:22:40,791 - hydromt.model.model - model - INFO - setup_other_demand.demand_fn=pcr_globwb
2026-01-14 11:22:40,791 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing water demand maps for ['industry_gross', 'industry_net', 'livestock_gross', 'livestock_net'].
2026-01-14 11:22:40,793 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading pcr_globwb RasterDataset data from /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/data/demand/pcr_globwb.nc
2026-01-14 11:22:40,976 - hydromt.model.model - model - INFO - update: setup_irrigation
2026-01-14 11:22:40,976 - hydromt.model.model - model - INFO - setup_irrigation.paddy_class=[12]
2026-01-14 11:22:40,976 - hydromt.model.model - model - INFO - setup_irrigation.area_threshold=0.6
2026-01-14 11:22:40,976 - hydromt.model.model - model - INFO - setup_irrigation.lai_threshold=0.2
2026-01-14 11:22:40,976 - hydromt.model.model - model - INFO - setup_irrigation.lulcmap_name=meta_landuse
2026-01-14 11:22:40,976 - hydromt.model.model - model - INFO - setup_irrigation.output_names={'irrigated_paddy_area__count': 'demand_paddy_irrigated_mask', 'irrigated_non_paddy_area__count': 'demand_nonpaddy_irrigated_mask', 'irrigated_paddy__irrigation_trigger_flag': 'demand_paddy_irrigation_trigger', 'irrigated_non_paddy__irrigation_trigger_flag': 'demand_nonpaddy_irrigation_trigger'}
2026-01-14 11:22:40,976 - hydromt.model.model - model - INFO - setup_irrigation.irrigated_area_fn=irrigated_area
2026-01-14 11:22:40,976 - hydromt.model.model - model - INFO - setup_irrigation.irrigation_value=[1]
2026-01-14 11:22:40,976 - hydromt.model.model - model - INFO - setup_irrigation.cropland_class=[11, 14, 20, 30]
2026-01-14 11:22:40,976 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing irrigation maps.
2026-01-14 11:22:40,977 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading irrigated_area RasterDataset data from /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/data/demand/irrigated_area.tif
2026-01-14 11:22:40,991 - hydromt.hydromt_wflow.workflows.demand - demand - INFO - Preparing irrigated areas map for non paddy.
2026-01-14 11:22:41,016 - hydromt.hydromt_wflow.workflows.demand - demand - INFO - Preparing irrigated areas map for paddy.
2026-01-14 11:22:41,041 - hydromt.hydromt_wflow.workflows.demand - demand - INFO - Calculating irrigation trigger.
2026-01-14 11:22:41,098 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Write model data to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand
2026-01-14 11:22:41,108 - hydromt.model.components.grid - grid - INFO - wflow_sbm.staticmaps: Writing grid data to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/staticmaps.nc.
2026-01-14 11:22:41,383 - hydromt.model.components.spatial - spatial - INFO - wflow_sbm.staticmaps: Writing region to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/staticgeoms/region.geojson.
2026-01-14 11:22:41,410 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/staticgeoms/region.geojson.
2026-01-14 11:22:41,411 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/staticgeoms/basins.geojson.
2026-01-14 11:22:41,414 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/staticgeoms/meta_basins_highres.geojson.
2026-01-14 11:22:41,428 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/staticgeoms/gauges_grdc.geojson.
2026-01-14 11:22:41,431 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/staticgeoms/outlets.geojson.
2026-01-14 11:22:41,432 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/staticgeoms/glaciers.geojson.
2026-01-14 11:22:41,453 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/staticgeoms/rivers.geojson.
2026-01-14 11:22:41,457 - hydromt.model.components.geoms - geoms - INFO - wflow_sbm.geoms: Writing geoms to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/staticgeoms/allocation_areas_admin2.geojson.
2026-01-14 11:22:41,461 - hydromt.hydromt_wflow.components.forcing - forcing - INFO - Write forcing file
2026-01-14 11:22:41,474 - hydromt.hydromt_wflow.components.forcing - forcing - INFO - Writing file /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/inmaps-era5-2010.nc
[########################################] | 100% Completed | 100.43 ms
2026-01-14 11:22:41,618 - hydromt.model.components.tables - tables - INFO - wflow_sbm.tables: No tables found, skip writing.
2026-01-14 11:22:41,618 - hydromt.model.components.grid - grid - INFO - wflow_sbm.states: No grid data found, skip writing.
2026-01-14 11:22:41,618 - hydromt.hydromt_wflow.components.config - config - INFO - Writing model config to /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/wflow_sbm.toml.
NOTE: The second catalog demand/data_catalog.yml points to additional data that are not available in the hydromt artifact_data catalog such as extracts of the glcnmo landuse or the pcrglobwb gridded demands.
Looking at the added layers#
To understand what layers have been added, we’ll plot the new layers below. To do the plotting, we first have to import the required python libraries:
[4]:
import numpy as np
import matplotlib.pyplot as plt
from hydromt_wflow import WflowSbmModel
# Open the wflow model
model = WflowSbmModel(
root="wflow_piave_water_demand",
mode="r+",
data_libs=[
"artifact_data",
"./data/demand/data_catalog.yml",
],
)
# Get the staticmaps
staticmaps = model.staticmaps.data
2026-01-14 11:22:44,430 - hydromt.data_catalog.data_catalog - data_catalog - INFO - Reading data catalog artifact_data latest
2026-01-14 11:22:44,431 - hydromt.data_catalog.data_catalog - data_catalog - INFO - Parsing data catalog from /home/runner/.hydromt/artifact_data/v1.0.0/data_catalog.yml
2026-01-14 11:22:45,070 - hydromt.data_catalog.data_catalog - data_catalog - INFO - Parsing data catalog from ./data/demand/data_catalog.yml
2026-01-14 11:22:45,092 - hydromt.model.model - model - INFO - Initializing wflow_sbm model from hydromt_wflow (v1.0.1.dev0).
2026-01-14 11:22:45,092 - hydromt.data_catalog.data_catalog - data_catalog - INFO - Parsing data catalog from /home/runner/work/hydromt_wflow/hydromt_wflow/hydromt_wflow/data/parameters_data.yml
2026-01-14 11:22:45,107 - hydromt.hydromt_wflow.wflow_base - wflow_base - INFO - Supported Wflow.jl version v1+
2026-01-14 11:22:45,108 - hydromt.hydromt_wflow.components.config - config - INFO - Reading model config file from /home/runner/work/hydromt_wflow/hydromt_wflow/docs/_examples/wflow_piave_water_demand/wflow_sbm.toml.
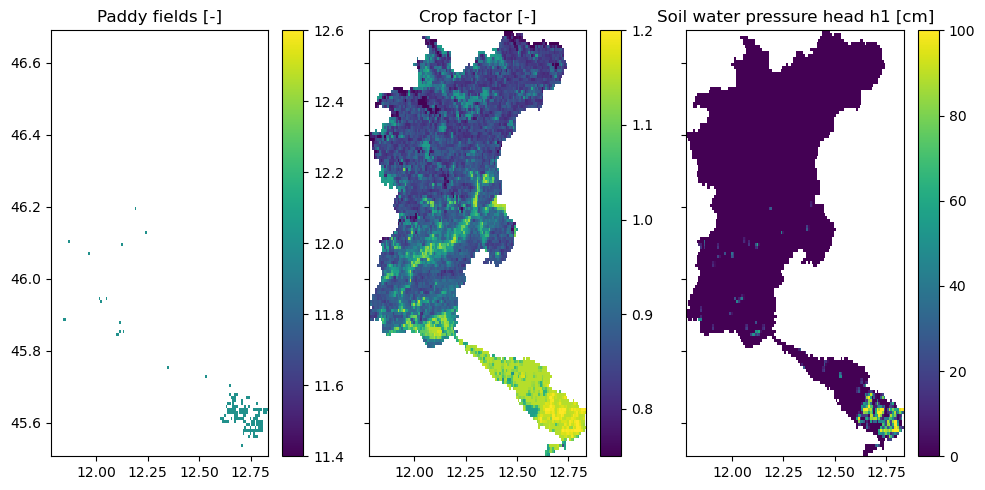
Landuse and rice fields#
Additional new parameters have been added to the model for landuse such as:
crop_factor: crop factor map that is used to convert the reference PET to the crop specific evaporation
h values: soil water pressure heads at which root water uptake is reduced (Feddes) [cm]. They are different for paddy and non paddy landuse types.
For paddies, new maps have been added and soil parameters updated to allow water to pool at the surface. These are the Brooks Corey c maps and a new soil_ksat_vertical_factor map which is a factor used to multiply the vertical conductivity (ksat vertical) in order to allow for a layer with very low conductivity (for the paddy fields).
[5]:
fig, axes = plt.subplots(1, 3, sharex=True, sharey=True, figsize=(10, 5))
axes[0].set_title("Paddy fields [-]")
axes[1].set_title("Crop factor [-]")
axes[2].set_title("Soil water pressure head h1 (Feddes) [cm]")
rice = staticmaps["meta_landuse"].where(
staticmaps["meta_landuse"] == 12, staticmaps["meta_landuse"].raster.nodata
)
rice.raster.mask_nodata().plot(ax=axes[0], add_labels=False)
staticmaps["vegetation_crop_factor"].raster.mask_nodata().plot(ax=axes[1], add_labels=False)
staticmaps["vegetation_feddes_h1"].raster.mask_nodata().plot(ax=axes[2], add_labels=False)
fig.tight_layout()
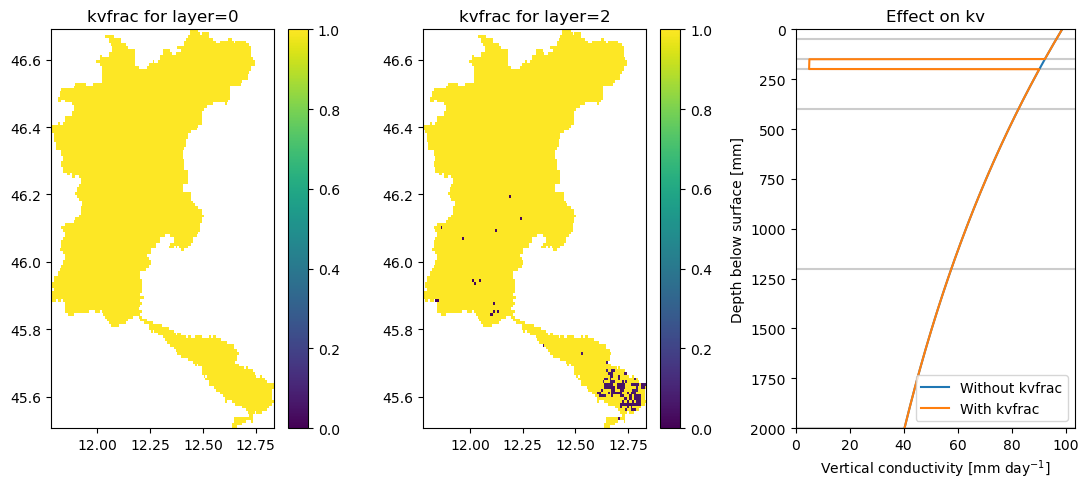
A soil_ksat_vertical_factor map is added that adds a layer with very low vertical hydraulic conductivity in the cells with paddy irrigation. The target conductivity value can be set for each layer using the target_conductivity parameter, and sets a value for each layer in the wflow model (linked to the wflow_thicknesslayer parameter). See the figures below for an indication of these maps and their effect on the vertical conductivity.
[6]:
fig, axes = plt.subplots(1, 3, figsize=(11, 5))
ax1, ax2, ax3 = axes.flatten()
ax1.set_title("soil_ksat_vertical_factor for layer=0")
ax2.set_title("soil_ksat_vertical_factor for layer=2")
ax3.set_title("Effect on kv")
staticmaps["soil_ksat_vertical_factor"].sel(layer=0).raster.mask_nodata().plot(
ax=ax1, add_labels=False, vmin=0, vmax=1
)
staticmaps["soil_ksat_vertical_factor"].sel(layer=2).raster.mask_nodata().plot(
ax=ax2, add_labels=False, vmin=0, vmax=1
)
# Take the cumulative sum to get to the cumulative layers
layers = np.cumsum(model.config.get_value("model.soil_layer__thickness"))
layers = np.append(layers, 2000)
# Add layers to figure
for layer in layers:
ax3.axhline(y=layer, c="0.8")
# Position of a paddy pixel
lat = 45.63
lon = 12.66
# Read required layers
kv_0 = staticmaps["soil_ksat_vertical"].sel(latitude=lat, longitude=lon, method="nearest")
soil_ksat_vertical_factor = staticmaps["soil_ksat_vertical_factor"].sel(latitude=lat, longitude=lon, method="nearest")
f = staticmaps["soil_f"].sel(latitude=lat, longitude=lon, method="nearest")
# Compute original kv values (without soil_ksat_vertical_factor)
depths = np.arange(0, 2000)
original = kv_0.values * np.exp(-f.values * depths)
# Compute new kv values
corrected = original.copy()
idxs = np.nonzero(soil_ksat_vertical_factor.values != 1)[0]
for idx in idxs:
start_depth = layers[idx - 1]
end_depth = layers[idx]
corrected[start_depth:end_depth] *= soil_ksat_vertical_factor.values[idx]
ax3.plot(original, depths, label="Without soil_ksat_vertical_factor")
ax3.plot(corrected, depths, label="With soil_ksat_vertical_factor")
# Flip y-axis for easier understanding of depth profile
ax3.set_ylim(2000, 0)
ax3.set_xlim(0, ax3.get_xlim()[1])
ax3.set_xlabel("Vertical conductivity [mm day$^{-1}$]")
ax3.set_ylabel("Depth below surface [mm]")
ax3.legend(loc="lower right")
fig.tight_layout()
Industry, livestock and domestic demand#
For the non_irrigation related demand, we assume those to be already prepared datasets with gross and net (consumption) demands. In this example, we rely on data from the PCR-GLOBWB model. See the images below for an explanation of the data and added layers to the wflow model configuration.
[7]:
month = 6
fig, axes = plt.subplots(2, 3, sharex=True, sharey=True, figsize=(10, 8))
fig.suptitle(f"Demand values for month {month}")
axes[0][0].set_title("Industry [mm/day]")
axes[0][1].set_title("Livestock [mm/day]")
axes[0][2].set_title("Domestic [mm/day]")
axes[0][0].set_ylabel("Gross")
axes[1][0].set_ylabel("Net")
# Extracting the min-max ranges for consistent colorbars
ind_min = staticmaps["demand_industry_gross"].min()
ind_max = staticmaps["demand_industry_gross"].max()
lsk_min = staticmaps["demand_livestock_gross"].min()
lsk_max = staticmaps["demand_livestock_gross"].max()
dom_min = staticmaps["demand_domestic_gross"].min()
dom_max = staticmaps["demand_domestic_gross"].max()
# Plot industry
staticmaps["demand_industry_gross"].sel(time=month).plot(
ax=axes[0][0], add_labels=False, vmin=ind_min, vmax=ind_max
)
staticmaps["demand_industry_net"].sel(time=month).plot(
ax=axes[1][0], add_labels=False, vmin=ind_min, vmax=ind_max
)
# Plot livestock
staticmaps["demand_livestock_gross"].sel(time=month).plot(
ax=axes[0][1], add_labels=False, vmin=lsk_min, vmax=lsk_max
)
staticmaps["demand_livestock_net"].sel(time=month).plot(
ax=axes[1][1], add_labels=False, vmin=lsk_min, vmax=lsk_max
)
# Plot domestic (adjusted the max range to improve plotting)
staticmaps["demand_domestic_gross"].sel(time=month).plot(
ax=axes[0][2], add_labels=False, vmin=dom_min, vmax=dom_max * 0.3
)
staticmaps["demand_domestic_net"].sel(time=month).plot(
ax=axes[1][2], add_labels=False, vmin=dom_min, vmax=dom_max * 0.3
)
fig.tight_layout()
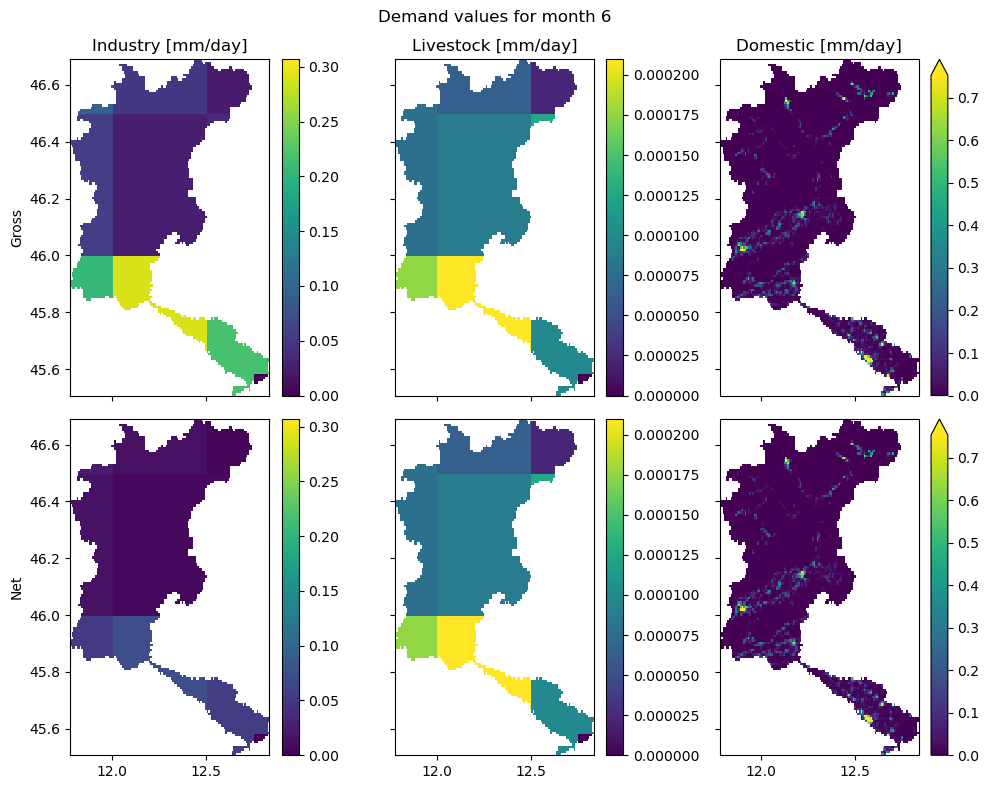
In the figure above, we see a slight difference between the gross and net demand for the industry sector. This means that this sector consumes part of the demanded water, but returns a portion of the water back. On the other hand, the gross and net demands for both livestock and domestic are roughly the same, meaning that the majority of the water will be consumed.
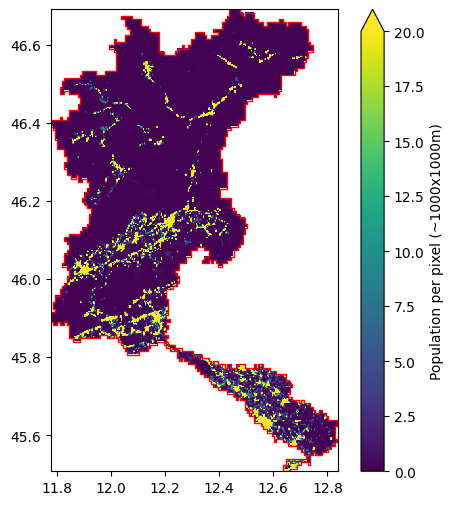
For the industry and livestock sectors, we see the relatively low-resolution of the original data. This data is retrieved from the PCR-GLOBWB model, which was provided at a resolution of 0.5x0.5 degrees (roughly 40x40km). The same holds for the domestic demands data, but it was downscaled using population density in the setup_non_irrigation workflow (using the dataset provided in population_fn, which is an optional step). The population data is used to identify densely populated regions,
and downscales this to the cities/villages where the people actually live. To give an impression, see the figure below for an insight into the population data. Please note that this is the data at the original resolution of ~100x100m. The data at the wflow model resolution is provided in the staticmaps (see mod.staticmaps.data["meta_population"]).
[8]:
# Read original data and slice to model domain
pop_ds = model.data_catalog.get_rasterdataset(
"ghs_pop_2015", geom=model.basins
)
# Get mask of catchment
pop_ds_mask = pop_ds.raster.geometry_mask(model.basins)
pop_ds = pop_ds.raster.mask_nodata().where(pop_ds_mask)
# Plot data
fig, ax = plt.subplots(1, 1, figsize=(6, 6))
# Add original population data to figure
pop_ds.plot(
ax=ax,
vmax=20,
add_labels=False,
cbar_kwargs={"label": "Population per pixel (~1000x1000m)"},
)
# Add basin geometry to plot
model.basins.plot(ax=ax, facecolor="none", edgecolor="red")
2026-01-14 11:22:47,254 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading ghs_pop_2015 RasterDataset data from /home/runner/.hydromt/artifact_data/latest/ghs_pop_2015.tif
[8]:
<Axes: >
Irrigation maps#
For the setup_irrigation workflow, a number of maps have been added to the model:
demand_paddy_irrigated_mask: mask (0/1) of the cells that are considered to be paddy/rice fields
demand_nonpaddy_irrigated_mask: mask (0/1) of the cells that are irrigated, but not paddy/rice fields
demand_nonpaddy_irrigation_trigger: trigger (0/1) that indicates whether we expect irrigation to occur (based on the LAI, to identify the growing season)
demand_paddy_irrigation_trigger: trigger (0/1) that indicates whether we expect irrigation to occur on paddies (based on the LAI, to identify the growing season)
[9]:
fig, axes = plt.subplots(1, 2, sharex=True, sharey=True, figsize=(10, 5))
axes[0].set_title("Paddy irrigation [-]")
axes[1].set_title("Non-paddy irrigation [-]")
staticmaps["demand_paddy_irrigated_mask"].raster.mask_nodata().plot(
ax=axes[0], add_labels=False
)
staticmaps["demand_nonpaddy_irrigated_mask"].raster.mask_nodata().plot(
ax=axes[1], add_labels=False
)
fig.tight_layout()
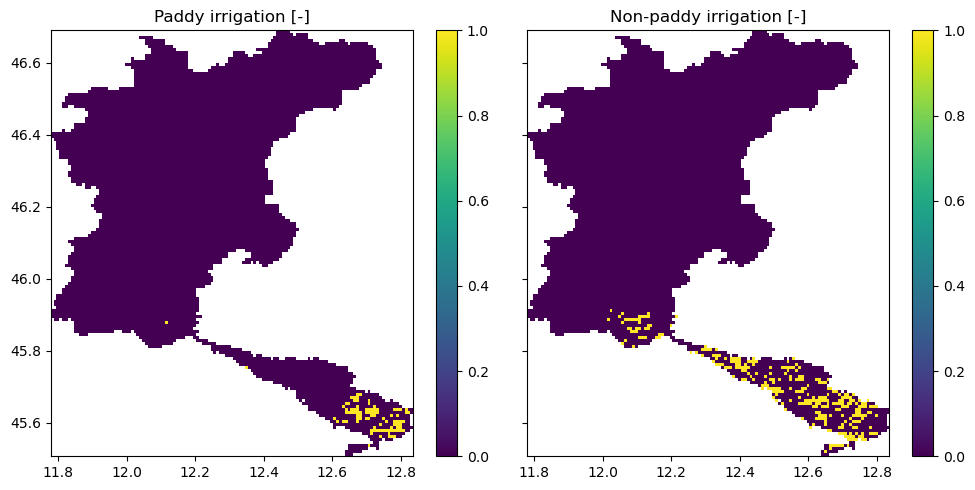
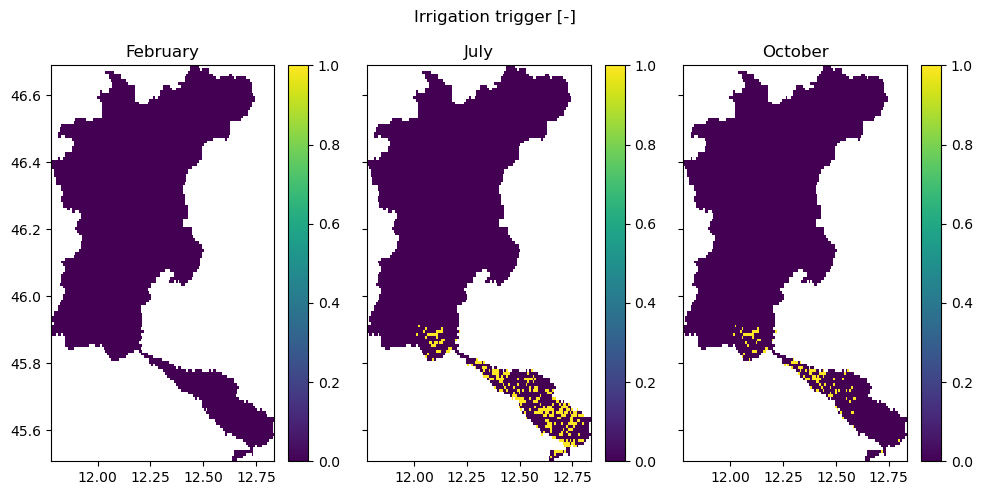
In this figure, we see the cells with paddy (rice fields) and non-paddy irrigated areas. Cells are identified as such when they exceed a fractional threshold of the cell (set by area_threshold). A cell can only be either paddy-irrigated, non-paddy-irrigated, or rain-fed. In the third panel, we see the crop factor associated for all of the model pixels.
In the figures below, we show the irrigation trigger maps for three months: before the growing season, during the growing season, and in the closing stages of the growing season. A map with a mask (0/1) contains information whether irrigation is allowed to occur during this month.
[10]:
fig, axes = plt.subplots(1, 3, sharex=True, sharey=True, figsize=(10, 5))
fig.suptitle("Irrigation trigger [-]")
axes[0].set_title("February")
axes[1].set_title("July")
axes[2].set_title("October")
staticmaps["demand_nonpaddy_irrigation_trigger"].sel(time=2).raster.mask_nodata().plot(
ax=axes[0], add_labels=False, vmin=0, vmax=1
)
staticmaps["demand_nonpaddy_irrigation_trigger"].sel(time=7).raster.mask_nodata().plot(
ax=axes[1], add_labels=False, vmin=0, vmax=1
)
staticmaps["demand_nonpaddy_irrigation_trigger"].sel(time=10).raster.mask_nodata().plot(
ax=axes[2], add_labels=False, vmin=0, vmax=1
)
fig.tight_layout()
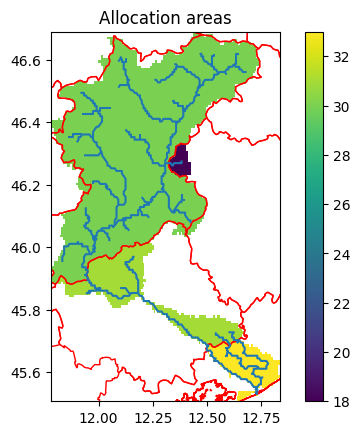
Water allocation regions#
To define regions where water can be shared and allocated, a merge between catchment and water areas or administrative boundaries is computed. These result in regions where wflow can allocate available water. No water allocation is supported between these regions. To give an impression on how these regions look like, see the following figures for an example.
Note: Water areas or regions are generally defined by sub-river-basins within a Country. In order to mimick reality, it is advisable to avoid cross-Country-border abstractions. Whenever information is available, it is strongly recommended to align the water regions with the actual areas managed by water management authorities, such as regional water boards.
[11]:
# Read original data and slice to model domain
admin = model.data_catalog.get_geodataframe("gadm_level2", geom=model.basins)
fig, ax = plt.subplots(1)
ax.set_title("Allocation areas")
staticmaps["allocation_areas_admin2"].raster.mask_nodata().plot(ax=ax, add_labels=False)
admin.plot(ax=ax, facecolor="none", edgecolor="red")
model.rivers.plot(ax=ax)
2026-01-14 11:22:48,469 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading gadm_level2 GeoDataFrame data from /home/runner/.hydromt/artifact_data/latest/gadm_level2.gpkg
[11]:
<Axes: title={'center': 'Allocation areas'}>
When merging the wflow basins with the water regions, some small subbasins can be created that do not contain river cells. These small basins will be merged to larger basins. When merging, you can decide if you prefer to merge with the nearest downstream basin, or with any basin in the same water region that does contain river using the priotity_basins argument. In the previous map, we gave priority to the basins, here is the results if priority is given to water regions instead:
[12]:
# Create allocations areas
model.setup_allocation_areas(
waterareas_fn="gadm_level2",
priority_basins=False,
)
fig, ax = plt.subplots(1)
ax.set_title("Allocation areas")
staticmaps["demand_allocation_area_id"].raster.mask_nodata().plot(
ax=ax, add_labels=False, cmap="viridis_r"
)
admin.plot(ax=ax, facecolor="none", edgecolor="red")
model.rivers.plot(ax=ax)
2026-01-14 11:22:48,715 - hydromt.hydromt_wflow.wflow_sbm - wflow_sbm - INFO - Preparing water demand allocation map.
2026-01-14 11:22:48,718 - hydromt.data_catalog.sources.data_source - data_source - INFO - Reading gadm_level2 GeoDataFrame data from /home/runner/.hydromt/artifact_data/latest/gadm_level2.gpkg
[12]:
<Axes: title={'center': 'Allocation areas'}>
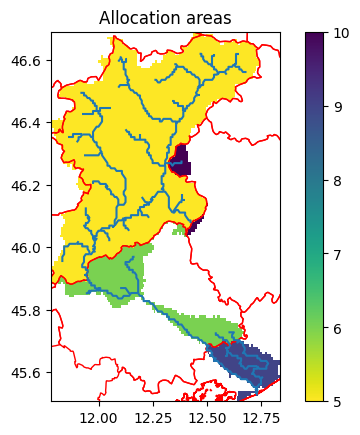
Here we see a couple of distinct administrative boundaries (black lines), some of which already follow the catchment boundaries. When the catchment crosses an administrative boundary, that region receives a different but unique identifier. Note that we are using level 2 boundaries here, which might not be the most realistic for a region of this size.
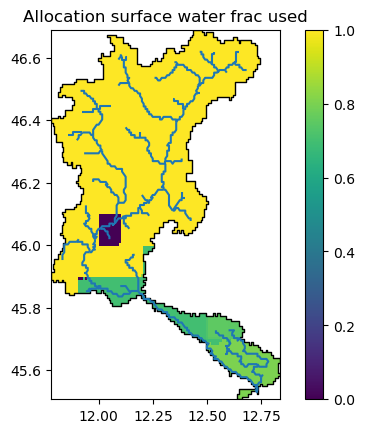
Surface water frac used for water allocation#
By default, Wflow will allocate all water demands with water from the surface water (demand_surface_water_ratio=1). However, if in a certain areas, groundwater or other non conventional sources can be used, the demand_surface_water_ratio of each allocation area can be reduced and prepared using the setup_allocation_surfacewaterfrac method.
Here we used global data from GLOFAS (Lisflood) for the fraction of grounwater used, presence of groundwater bodies and non conventional sources (0 is Piave). The water allocations are the ones we prepared in the previous step in order to match better the wflow model basin delineation.
Let’s have a look at the resulting map:
[13]:
fig, ax = plt.subplots(1)
ax.set_title("Allocation surface water frac used")
staticmaps["demand_surface_water_ratio"].raster.mask_nodata().plot(ax=ax, add_labels=False)
model.basins.plot(ax=ax, facecolor="none", edgecolor="black")
model.rivers.plot(ax=ax)
[13]:
<Axes: title={'center': 'Allocation surface water frac used'}>