Example: Reading geospatial point time-series#
This example illustrates the how to read geospatial point time-series data here called GeoDataset, using the HydroMT DataCatalog with the vector and netcdf or zarr drivers.
[1]:
# import hydromt and setup logging
import hydromt
from hydromt.log import setuplog
import numpy as np
from pprint import pprint
logger = setuplog("read geodataset data", log_level=10)
2024-02-26 15:12:00,923 - read geodataset data - log - INFO - HydroMT version: 0.9.4
[2]:
# Download artifacts for the Piave basin to `~/.hydromt_data/`.
data_catalog = hydromt.DataCatalog(logger=logger, data_libs=["artifact_data"])
2024-02-26 15:12:00,954 - read geodataset data - data_catalog - INFO - Reading data catalog archive artifact_data v0.0.8
2024-02-26 15:12:00,955 - read geodataset data - data_catalog - INFO - Parsing data catalog from /home/runner/.hydromt_data/artifact_data/v0.0.8/data_catalog.yml
Netcdf or zarr driver#
To read geospatial point time-series data and parse it into a xarray Dataset or DataArray we use the open_mfdataset() or the open_zarr() method. All driver_kwargs in the data catalog yaml file will be passed to to these methods.
As an example we will use the GTSM data dataset which is stored in Netcdf format.
[3]:
# inspect data source entry in data catalog yaml file
data_catalog["gtsmv3_eu_era5"]
[3]:
crs: 4326
data_type: GeoDataset
driver: netcdf
meta:
category: ocean
paper_doi: 10.24381/cds.8c59054f
paper_ref: Copernicus Climate Change Service 2019
source_license: https://cds.climate.copernicus.eu/cdsapp/#!/terms/licence-to-use-copernicus-products
source_url: https://cds.climate.copernicus.eu/cdsapp#!/dataset/10.24381/cds.8c59054f?tab=overview
source_version: GTSM v3.0
path: /home/runner/.hydromt_data/artifact_data/v0.0.8/gtsmv3_eu_era5.nc
We can load any geospatial point time-series data using DataCatalog.get_geodataset(). Note that if we don’t provide any arguments it returns the full dataset with all data variables and for the full spatial domain. The result is per default returned as DataArray as the dataset consists of a single variable. To return a dataset use the single_var_as_array=False argument. Only the
data coordinates and the time are actually read, the data variables are still lazy Dask arrays.
[4]:
ds = data_catalog.get_geodataset("gtsmv3_eu_era5", single_var_as_array=False)
ds
[4]:
<xarray.Dataset> Size: 323kB
Dimensions: (stations: 19, time: 2016)
Coordinates:
lon (stations) float64 152B dask.array<chunksize=(19,), meta=np.ndarray>
* time (time) datetime64[ns] 16kB 2010-02-01 ... 2010-02-14T23:50:00
lat (stations) float64 152B dask.array<chunksize=(19,), meta=np.ndarray>
spatial_ref int32 4B ...
* stations (stations) int32 76B 13670 2798 2799 13775 ... 2791 2790 2789
Data variables:
waterlevel (time, stations) float64 306kB dask.array<chunksize=(2016, 19), meta=np.ndarray>
Attributes:
category: ocean
paper_doi: 10.24381/cds.8c59054f
paper_ref: Copernicus Climate Change Service 2019
source_license: https://cds.climate.copernicus.eu/cdsapp/#!/terms/licenc...
source_url: https://cds.climate.copernicus.eu/cdsapp#!/dataset/10.24...
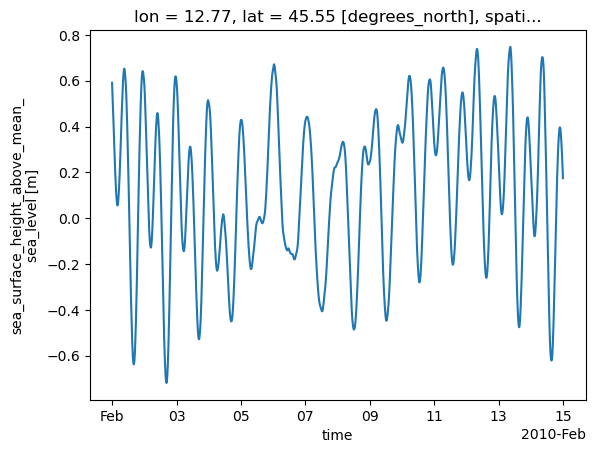
source_version: GTSM v3.0The data can be visualized with the .plot() xarray method. We show the evolution of the water level over time for a specific point location (station).
[5]:
ds.sel(stations=2791)["waterlevel"].plot()
[5]:
[<matplotlib.lines.Line2D at 0x7f0d19d2d690>]
We can request a (spatial) subset data by providing additional variables and bbox / geom arguments. Note that these return less stations and less variables. In this example only spatial arguments are applied as only a single variable is available. The variables argument is especially useful if each variable of the dataset is saved in a separate file and the {variable} key is used in the path argument of the data source to limit which files are actually read. If a single variable
is requested a DataArray instead of a Dataset is returned unless the single_var_as_array argument is set to False (True by default).
[6]:
bbox = [12.50, 45.20, 12.80, 45.40]
ds_bbox = data_catalog.get_geodataset("gtsmv3_eu_era5", bbox=bbox)
ds_bbox
[6]:
<xarray.DataArray 'waterlevel' (time: 2016, stations: 2)> Size: 32kB
dask.array<getitem, shape=(2016, 2), dtype=float64, chunksize=(2016, 2), chunktype=numpy.ndarray>
Coordinates:
lon (stations) float64 16B dask.array<chunksize=(2,), meta=np.ndarray>
* time (time) datetime64[ns] 16kB 2010-02-01 ... 2010-02-14T23:50:00
lat (stations) float64 16B dask.array<chunksize=(2,), meta=np.ndarray>
spatial_ref int32 4B ...
* stations (stations) int32 8B 13775 13723
Attributes:
long_name: sea_surface_height_above_mean_sea_level
units: m
CDI_grid_type: unstructured
short_name: waterlevel
description: Total water level resulting from the combination of baro...
category: ocean
paper_doi: 10.24381/cds.8c59054f
paper_ref: Copernicus Climate Change Service 2019
source_license: https://cds.climate.copernicus.eu/cdsapp/#!/terms/licenc...
source_url: https://cds.climate.copernicus.eu/cdsapp#!/dataset/10.24...
source_version: GTSM v3.0With a Geodataset, you can also directly access the associated point geometries using its to_gdf method. Multi-dimensional data (e.g. the time series) can be reduced using a statistical method such as the max.
[7]:
pprint(ds_bbox.vector.to_gdf(reducer=np.nanmax))
geometry waterlevel
stations
13775 POINT (12.75879 45.24902) 0.703679
13723 POINT (12.50244 45.25635) 0.721433
[8]:
# Plot
import matplotlib.pyplot as plt
import geopandas as gpd
from shapely.geometry import box
import cartopy.io.img_tiles as cimgt
import cartopy.crs as ccrs
proj = ccrs.PlateCarree()
fig = plt.figure(figsize=(6, 10))
ax = plt.subplot(projection=proj)
bbox = gpd.GeoDataFrame(geometry=[box(12.50, 45.20, 12.80, 45.40)], crs=4326)
ax.add_image(cimgt.QuadtreeTiles(), 12)
# Plot the points
ds.vector.to_gdf().plot(ax=ax, markersize=30, c="blue", zorder=2)
ds_bbox.vector.to_gdf().plot(ax=ax, markersize=40, c="red", zorder=2)
# Plot the bounding box
bbox.boundary.plot(ax=ax, color="red", linewidth=0.8)
[8]:
<GeoAxes: >
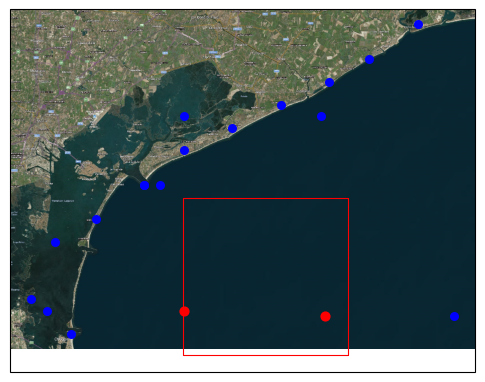
Vector driver#
To read vector data and parse it into a xarray.Dataset object we use the open_geodataset method. Combined point locations (e.g. CSV or GeoJSON) data as well as text delimited time series (e.g. CSV) data are supported as file formats (see DataCatalog documentation). Both formats must contain an index and a crs has to be indicated in the .yml file. For demonstration we use dummy example data from the examples/data folder.
First load the data catalog of the corresponding example data geodataset_catalog.yml:
[9]:
import os
geodata_catalog = hydromt.DataCatalog("data/geodataset_catalog.yml", logger=logger)
geodata_catalog["waterlevels_txt"]
2024-02-26 15:12:05,208 - read geodataset data - data_catalog - INFO - Parsing data catalog from data/geodataset_catalog.yml
[9]:
crs: 4326
data_type: GeoDataset
driver: vector
driver_kwargs:
fn_data: /home/runner/work/hydromt/hydromt/docs/_examples/data/stations_data.csv
path: /home/runner/work/hydromt/hydromt/docs/_examples/data/stations.csv
rename:
stations_data: waterlevel
We see here that our locations are defined in a data/stations.csv file and a data/stations_data.csv. Let’s check the content of these files before loading them with the get_geodataset method:
[10]:
import pandas as pd
print("Stations locations:")
df_stations = pd.read_csv("data/stations.csv")
pprint(df_stations)
Stations locations:
stations x y
0 1001 12.50244 45.25635
1 1002 12.75879 45.24902
[11]:
print("Stations data:")
df_stations_data = pd.read_csv("data/stations_data.csv")
pprint(df_stations_data.head(3))
Stations data:
time 1001 1002
0 2016-01-01 0.590860 0.591380
1 2016-01-02 0.565552 0.571342
2 2016-01-03 0.538679 0.549770
[12]:
ds = geodata_catalog.get_geodataset("waterlevels_txt", single_var_as_array=False)
ds
[12]:
<xarray.Dataset> Size: 48kB
Dimensions: (time: 2016, stations: 2)
Coordinates:
* time (time) datetime64[ns] 16kB 2016-01-01 2016-01-02 ... 2021-07-08
* stations (stations) int64 16B 1001 1002
geometry (stations) object 16B dask.array<chunksize=(2,), meta=np.ndarray>
spatial_ref int64 8B 0
Data variables:
waterlevel (stations, time) float64 32kB dask.array<chunksize=(2, 2016), meta=np.ndarray>