Note
Go to the end to download the full example code.
Quick overview#
Here are a number of quick examples of how to get started with xugrid. More detailed explanation can be found in the rest of the documentation.
We’ll start by importing a few essential packages.
import numpy as np
import xarray as xr
import xugrid as xu
Create a UgridDataArray#
There are three ways to create a UgridDataArray:
From an xarray Dataset containing the grid topology stored according to the UGRID conventions.
From a xugrid Ugrid object and an xarray DataArray containing the data.
From a UGRID netCDF file, via
xugrid.open_dataset().
From xarray Dataset#
xugrid will automatically find the UGRID topological variables, and separate them from the main data variables.
Details on the required variables can be found in the UGRID conventions. For 1D and 2D UGRID topologies, the required variables are:
x-coordinates of the nodes
y-coordinates of the nodes
edge node connectivity (1D) or face node connectivity (2D)
a “dummy” variable storing the names of the above variables in its attributes
We’ll start by fetching a dataset:
ds = xu.data.adh_san_diego(xarray=True)
ds
There are a number of topology coordinates and variables: node_x and
node_y, mesh2d and face_node_connectivity. The dummy variable
is mesh2d contains only a 0 for data; its attributes contain a mapping of
UGRID roles to dataset variables.
We can convert this dataset to a UgridDataset which will automatically separate the variables:
uds = xu.UgridDataset(ds)
uds
We can then grab one of the data variables as usual for xarray:
elev = uds["elevation"]
elev
From Ugrid and DataArray#
Alternatively, we can build a Ugrid topology object first from vertices and connectivity numpy arrays, for example when using the topology data generated by a mesh generator (at which stage there is no data asssociated with the nodes, edges, or faces).
There are many ways to construct such arrays, typically via mesh generators or Delaunay triangulation, but we will construct two simple triangles and some data by hand here:
nodes = np.array([[0, 0], [0, 1.1], [1, 0], [1, 1]])
faces = np.array([[2, 3, 0], [3, 1, 0]])
fill_value = -1
grid = xu.Ugrid2d(nodes[:, 0], nodes[:, 1], fill_value, faces)
da = xr.DataArray(
data=[1.0, 2.0],
dims=[grid.face_dimension],
)
uda = xu.UgridDataArray(da, grid)
uda
From netCDF file#
xugrid.open_dataset() is demonstrated in the last section of this
guide. Internally, it opens the netCDF as a regular dataset, then converts it
as seen in the first example.
Plotting#
elev.ugrid.plot(cmap="viridis")
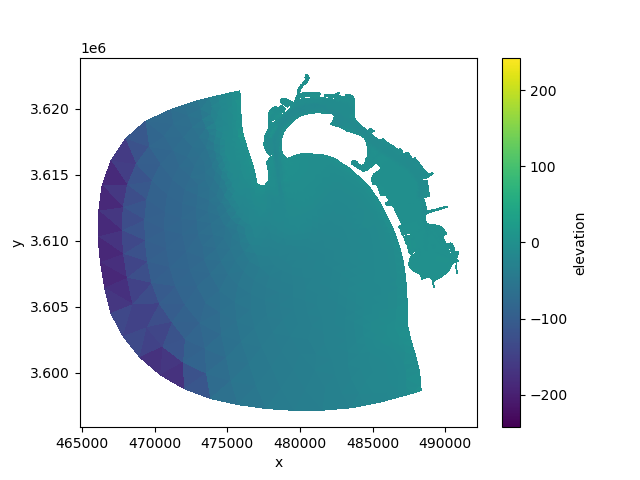
<matplotlib.collections.PolyCollection object at 0x7fb384a37bc0>
Data selection#
A UgridDataArray behaves identical to an xarray DataArray:
whole = xu.data.disk()["face_z"]
To select based on the topology, use the .ugrid attribute:
subset = whole.ugrid.sel(y=slice(5.0, None))
subset.ugrid.plot()
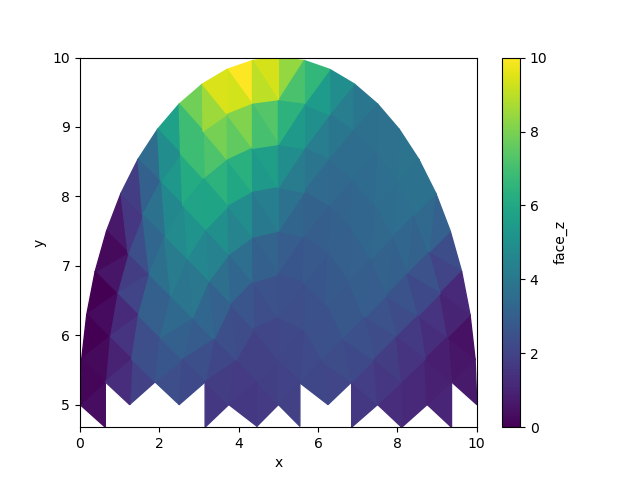
<matplotlib.collections.PolyCollection object at 0x7fb383b494f0>
Note
ugrid.sel() currently only supports data on the faces for 2D
topologies, and data on edges for 1D topologies. More flexibility
may be added.
Computation#
Computation on DataArrays is unchanged from xarray:
uda + 10.0
Geopandas#
Xugrid objects provide a number of conversion functions from and to geopandas
GeoDataFrames using xugrid.UgridDataset.from_geodataframe(). Note
that storing large grids as GeoDataFrames can be very inefficient.
gdf = uda.ugrid.to_geodataframe(name="test")
gdf
Conversion from Geopandas is easy too:
xu.UgridDataset.from_geodataframe(gdf)
XugridDatasets#
Like an Xarray Dataset, a UgridDataset is a dict-like container of UgridDataArrays. It is required that they share the same grid topology; but the individual DataArrays may be located on different aspects of the grid (nodes, faces, edges).
xu.data.disk()
A UgridDataset may be initialized without data variables, but this requires a grid object:
new_uds = xu.UgridDataset(grids=uds.ugrid.grids)
new_uds
We can then add variables one-by-one, as we might with an xarray Dataset:
new_uds["elevation"] = elev
new_uds
Write netCDF files#
Once again like xarray, NetCDF is the recommended file format for xugrid objects. Xugrid automatically stores the grid topology according to the UGRID conventions and merges it with the main dataset containing the data variables before writing.
uds.ugrid.to_netcdf("example-ugrid.nc")
xu.open_dataset("example-ugrid.nc")
/home/runner/work/xugrid/xugrid/xugrid/ugrid/ugrid2d.py:404: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
dataset = dataset.merge(other)
/home/runner/work/xugrid/xugrid/xugrid/ugrid/ugrid2d.py:404: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
dataset = dataset.merge(other)
Total running time of the script: (0 minutes 0.541 seconds)