Note
Go to the end to download the full example code.
Partitioning#
Grid partitioning, or domain decomposition, is an important step in setting up parallellized simulations. Xugrid provides utilities for partitioning a grid and its associated data, and for merging partitions back into a single whole.
import matplotlib.pyplot as plt
import numpy as np
import xugrid as xu
Create partitions#
Xugrid wraps the well known METIS library via the pymetis bindings. METIS is generally used to partition a grid in such a manner that communication between parallel processes is minimized.
We’ll demonstrate the functionality by diving the elevation example into several parts.
uda = xu.data.elevation_nl()
uda.ugrid.plot(vmin=-20, vmax=90, cmap="terrain")
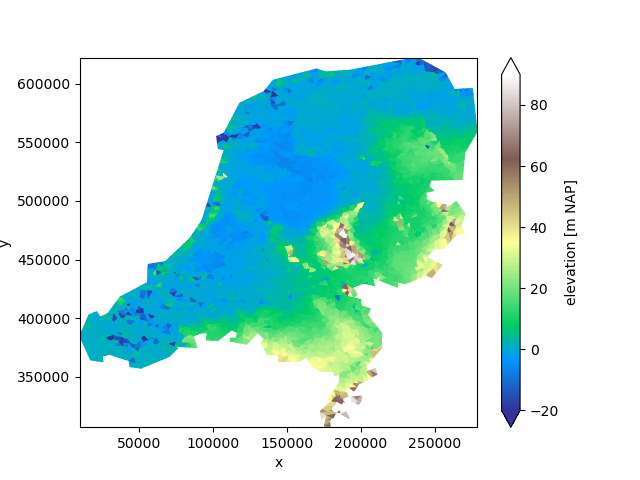
<matplotlib.collections.PolyCollection object at 0x7fb33b629100>
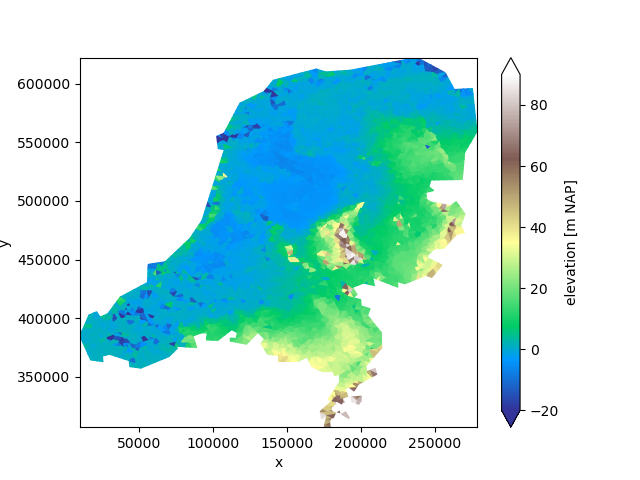
Let’s start by dividing the grid into four parts:
partitions = uda.ugrid.partition(n_part=4)
fig, axes = plt.subplots(2, 2, figsize=(12.6, 10))
for partition, ax in zip(partitions, axes.ravel()):
partition.ugrid.plot(ax=ax, vmin=-20, vmax=90, cmap="terrain")
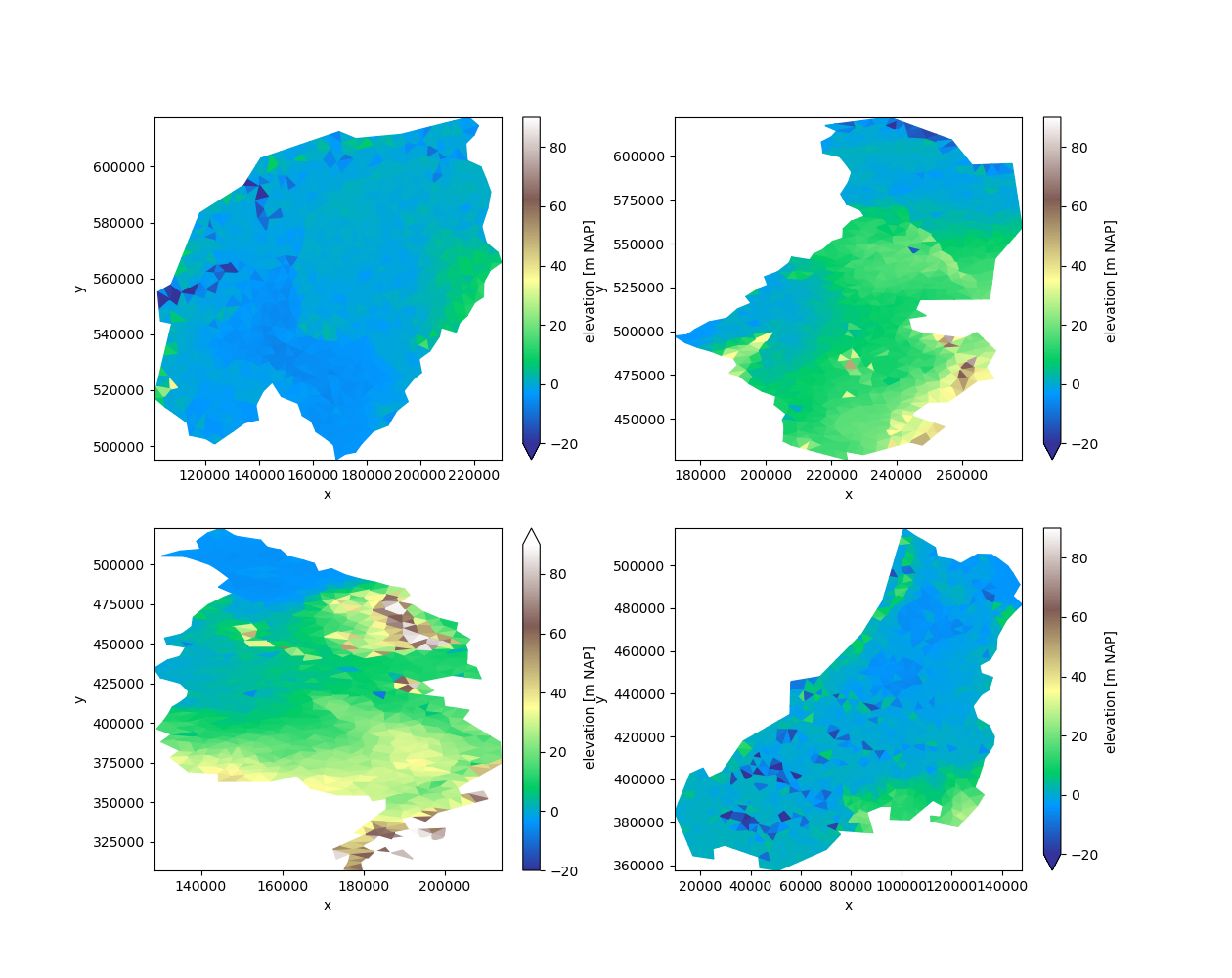
Partition the grid#
Calling .partition on a UgridDataArray or UgridDataset will automatically
partition the grid topology, select all associated data, and create a new
UgridDataArray or UgridDataset for each partition.
However, in some case, we might prefer to pre-compute the labels, and then
apply them multiple datasets. To do so, we compute the partition labels from
the grid. label_partitions returns a UgridDataArray, with every cell given
its partition label number.
We can easily plot this data to visualize the partitions:
labels = uda.ugrid.grid.label_partitions(n_part=12)
labels.ugrid.plot()
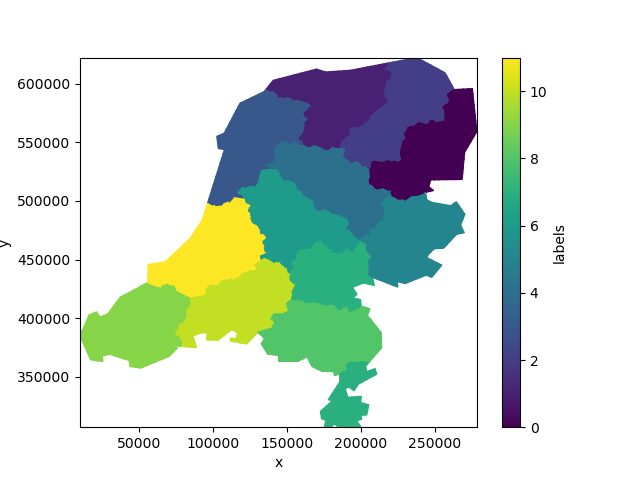
<matplotlib.collections.PolyCollection object at 0x7fb3848d7b90>
Not quite the twelve provinces of the Netherlands!
However, we may use the labels to partition the data nonetheless:
partitions = uda.ugrid.partition_by_label(labels)
fig, axes = plt.subplots(4, 3, figsize=(15, 15))
for partition, ax in zip(partitions, axes.ravel()):
partition.ugrid.plot(ax=ax, vmin=-20, vmax=90, cmap="terrain")
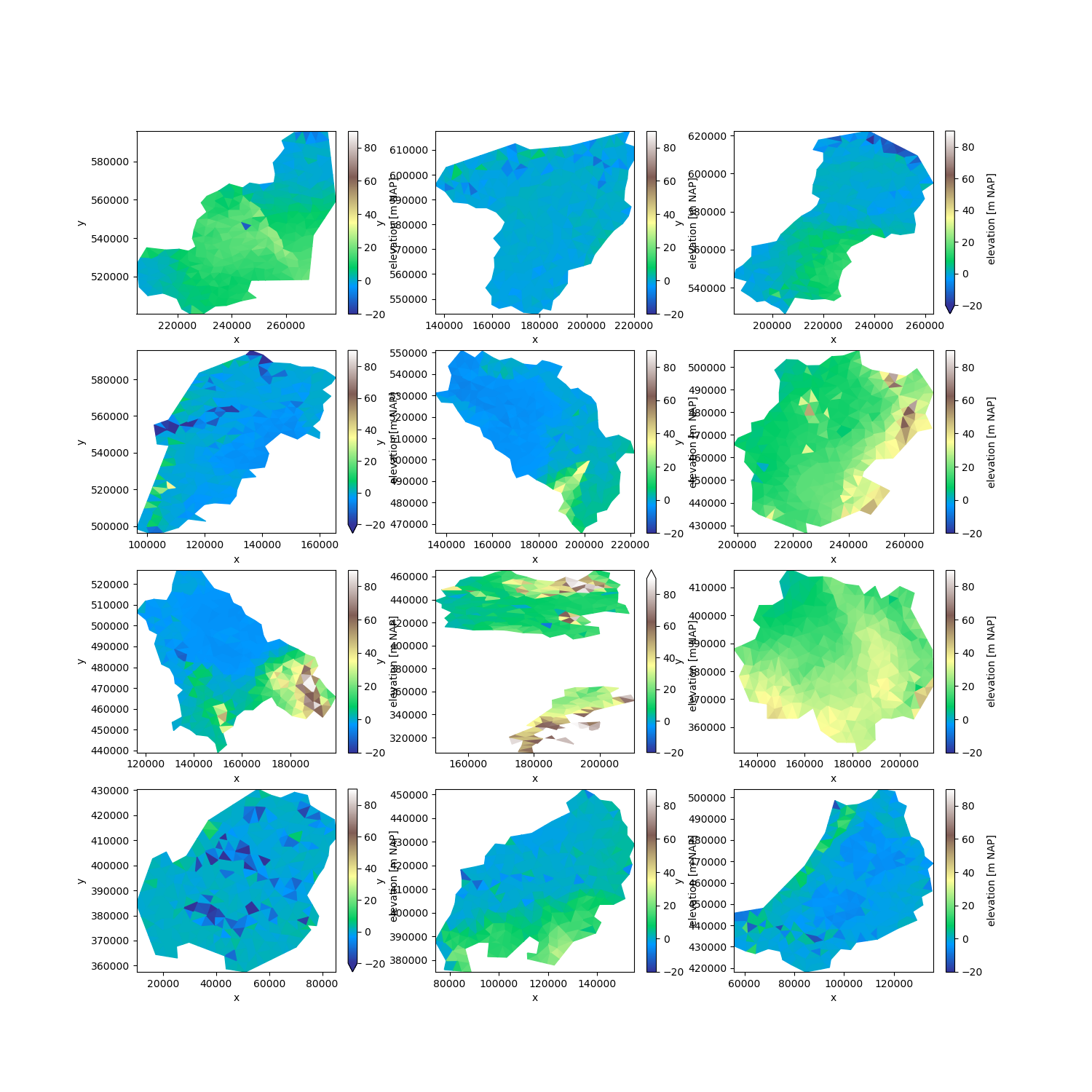
Since the labels are an ordinary UgridDataArray, we can easily store them in a netCDF file and re-use them in another part of a workflow.
Merging partitions#
Generally, after partitioning the data we write it as model input and run a model in parallel. Many model codes produce output per process. Xugrid can merge these partitions back into one whole for post-processing:
merged = xu.merge_partitions(partitions)["elevation"]
merged.ugrid.plot(vmin=-20, vmax=90, cmap="terrain")
<matplotlib.collections.PolyCollection object at 0x7fb383be6fc0>
Partitioning grids without data#
Of course, we can also partition the grid topology without any associated data:
grid = uda.ugrid.grid
grid_parts = grid.partition(n_part=4)
fig, axes = plt.subplots(2, 2, figsize=(12.6, 10))
for part, ax in zip(grid_parts, axes.ravel()):
part.plot(ax=ax)
… and merge them back into one whole:
merged_grid, _ = xu.Ugrid2d.merge_partitions(grid_parts)
merged_grid.plot()
<matplotlib.collections.LineCollection object at 0x7fb339fbfce0>
Preserving order#
Note that partioning and merging does not preserve order!
uda == merged
The topology is equivalent, but the nodes, edges, and faces are in a
different order. This is because merge_partitions simply concatenates the
partitions.
The easiest way to restore the order is by providing an example of the
original topology. reindex_like looks at the coordinates of both
(equivalent!) grids and automatically determines how to reorder:
reordered = merged.ugrid.reindex_like(uda)
uda == reordered
Alternatively, we can also assign IDs, carry these along, and use these to reorder the data after merging.
uds = xu.UgridDataset(grids=[uda.ugrid.grid])
uds["elevation"] = uda
uds["cell_id"] = ("mesh2d_nFaces", np.arange(len(uda)))
partitions = uds.ugrid.partition(n_part=4)
merged = xu.merge_partitions(partitions)
order = np.argsort(merged["cell_id"].values)
reordered = merged.isel(mesh2d_nFaces=order)
uds["elevation"] == reordered["elevation"]
This is required if results are compared with the input, or with results stemming from another partitioning, e.g. one with a different number of partitions.
Total running time of the script: (0 minutes 3.287 seconds)