Modelbuilder#
This notebook presents the basic features of the modelbuilder proof of concept. It generates a D-Flow FM model from scratch with only spatial and temporal extents as input. Since this is a proof of concept, many functions/inputs will change in the future and this notebook will be updated accordingly.
This modelbuilder combines many other Python packages. We use MeshKernelPy for grid generation, HYDROLIB-core to write all D-Flow FM files (like .mdu, .ext, *.bc), xarray to process netcdf files and xugrid to process ugrid netcdf files. Everything is wrapped in dfm_tools which also provide additional features.
If you prefer to interact with a Python script instead, you can convert this notebook to *.py with jupyter nbconvert --to script modelbuilder_example.ipynb.
Exercises#
Some of the blocks in in this notebook contain exercises. It is recommended to first just run the entire notebook without doing the exercises to understand the modelbuilding process. If you want to discover more, you can use these exercises as a first step.
1. Registering for data retrieval#
In this notebook we use publicly available data from Copernicus Programme of the European Union. To access this data you need to create accounts at Copernicus Marine Service and the Climate Data Store. Do not forget to accept the CDS license agreement. You will be promted for your CMEMS credentials and CDS apikey by this notebook automatically and they will be stored locally. If you want to avoid the prompt, set the environment variables COPERNICUSMARINE_SERVICE_USERNAME, COPERNICUSMARINE_SERVICE_PASSWORD and CDSAPI_KEY.
2. Imports and user variables#
We start by importing the libraries that are used in this notebook and setting some user input variables that for instance define the spatial and time extent of the resulting model.
Exercises#
change the model simulation period (
date_minanddate_maxvariables)create a new
model_nameand corresponding domain (lon_min,lon_max,lat_min,lat_maxvariables). Use an area of approximately 1x1 degrees for a quick test.
[1]:
# import packages
import os
from pathlib import Path
import matplotlib.pyplot as plt
plt.close('all')
import dfm_tools as dfmt
import hydrolib.core.dflowfm as hcdfm
import xarray as xr
import pandas as pd
import numpy as np
import contextily as ctx
[2]:
# user input
model_name = 'Vietnam' # the name cannot contain a space
overwrite = False # overwrite the downloaded forcing data or not. Always set to True when changing the domain
crs = 'EPSG:4326' # coordinate reference system, only EPSG 4326 (WGS84) is currently supported by the ERA5 and CMEMS download scripts.
modeltype = "2D" # set to "3D" for a 3D model including salinity and temperature
dir_output = os.path.abspath(f'./{model_name}_{modeltype}_model')
# path_style = 'windows' # windows / unix
# domain and resolution
# the actual maximum extents can slightly vary: see dfmt.meshkernel_get_bbox() below
if model_name=='Bonaire':
lon_min, lon_max, lat_min, lat_max = -68.55, -67.9, 11.8, 12.6
elif model_name=='Vietnam':
lon_min, lon_max, lat_min, lat_max = 105.8, 106.85, 17.75, 18.5
dxy = 0.05
# dates as understood by pandas.period_range(). ERA5 has freq='M' (month) and CMEMS has freq='D' (day)
date_min = '2022-11-01'
date_max = '2022-11-03'
ref_date = '2022-01-01'
[3]:
# make directories for the model and the downloaded data
os.makedirs(dir_output, exist_ok=True)
dir_output_data = os.path.join(dir_output, 'data')
os.makedirs(dir_output_data, exist_ok=True)
3. Grid generation and refinement with meshkernelpy#
To build a model we first need to generate a grid. We do this with MeshKernelPy, a Python wrapper for the MeshKernel, which is the Deltares C++ library for creating and editing meshes. It supports 1D & 2D unstructured meshes as well as curvilinear meshes. We will create a 2D unstructured mesh (grid) in this example.
We start by generating a basegrid for the spatial extent and generate a polygon for the seaward boundary. This is followed by refinement based on GEBCO bathymetry, cutting away the landward part of the grid. Lastly, we interpolate bathymetry to the grid and save the grid as a network file (*_net.nc file). The seaward boundary is derived from the grid extents and converted to a HYDROLIB-core dflowfm.PolyFile (*.pli file). With these files, the
first part of the D-Flow FM model is generated.
More advanced examples of grid generation are available at the MeshkernelPy Github and the dfm_tools Github.
Exercises#
apply more/less refinement to the grid (
min_edge_sizevariable)generate a
PolyFilewith more/less points (resvariable indfmt.interpolate_bndpli())
[4]:
# generate spherical regular grid
mk_object = dfmt.make_basegrid(lon_min=lon_min, lon_max=lon_max, lat_min=lat_min, lat_max=lat_max, dx=dxy, dy=dxy, crs=crs)
# retrieve actual lat/lon bounds from grid, the lon_max and lat_max are likely larger than requested
lon_min, lat_min, lon_max, lat_max = dfmt.meshkernel_get_bbox(mk_object)
# plot basegrid and polyline
fig, ax = plt.subplots()
mk_object.mesh2d_get().plot_edges(ax, zorder=1)
# ctx.add_basemap(ax=ax, crs=crs, attribution=False)
dfmt.plot_coastlines(ax=ax, crs=crs)
>> reading coastlines: 4.06 sec
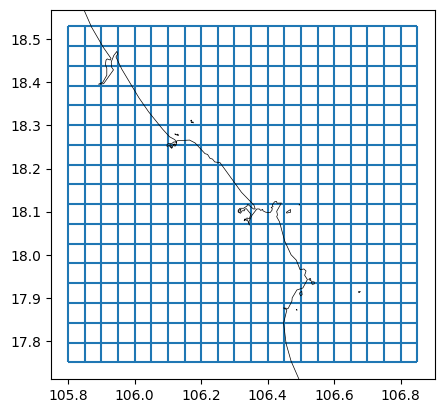
[5]:
# generate plifile from grid extent and coastlines
bnd_gdf = dfmt.generate_bndpli_cutland(mk=mk_object, res='h', buffer=0.01)
bnd_gdf_interp = dfmt.interpolate_bndpli(bnd_gdf, res=0.03)
pli_polyfile = dfmt.geodataframe_to_PolyFile(bnd_gdf_interp, name=f'{model_name}_bnd')
poly_file = os.path.join(dir_output, f'{model_name}.pli')
pli_polyfile.save(poly_file)
# plot basegrid and polyline
fig, ax = plt.subplots()
mk_object.mesh2d_get().plot_edges(ax, zorder=1, edgecolor='grey')
bnd_gdf_interp.plot(ax=ax, edgecolor='r')
# ctx.add_basemap(ax=ax, crs=crs, attribution=False)
dfmt.plot_coastlines(ax=ax, crs=crs)
>> reading coastlines: 2.15 sec
>> reading coastlines: 4.29 sec
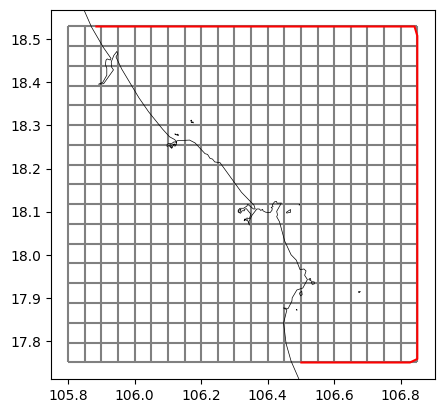
[6]:
# connect to a coarse version of the GEBCO_2022 dataset on OPeNDAP
file_nc_bathy = "https://opendap.deltares.nl/thredds/dodsC/opendap/deltares/Delft3D/netcdf_example_files/GEBCO_2022/GEBCO_2022_coarsefac08.nc"
data_bathy = xr.open_dataset(file_nc_bathy).elevation
# alternatively download your own full resolution cutout from https://download.gebco.net (use a buffer of e.g. 1 degree), also available at
# file_nc_bathy = r'p:\metocean-data\open\GEBCO\2022\GEBCO_2022.nc'
# data_bathy = xr.open_dataset(file_nc_bathy).elevation
# alternatively you can connect to ETOPO 30s, for which there is also a 15s (15 arcseconds) resolution dataset available
# file_nc_bathy = "https://www.ngdc.noaa.gov/thredds/dodsC/global/ETOPO2022/30s/30s_surface_elev_netcdf/ETOPO_2022_v1_30s_N90W180_surface.nc"
# data_bathy = xr.open_dataset(file_nc_bathy).z
# subset to area of interest
data_bathy_sel = data_bathy.sel(lon=slice(lon_min-1, lon_max+1), lat=slice(lat_min-1, lat_max+1))
# refine grid (Courant refinement)
min_edge_size = 300 # in meters
dfmt.refine_basegrid(mk=mk_object, data_bathy_sel=data_bathy_sel, min_edge_size=min_edge_size)
# plot
fig, ax = plt.subplots()
mk_object.mesh2d_get().plot_edges(ax,zorder=1)
# ctx.add_basemap(ax=ax, crs=crs, attribution=False)
dfmt.plot_coastlines(ax=ax, crs=crs)
>> reading coastlines: 2.40 sec
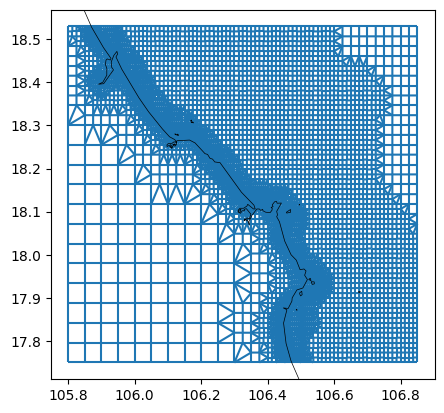
[7]:
# remove land with GSHHS coastlines
dfmt.meshkernel_delete_withcoastlines(mk=mk_object, res='h')
# derive illegalcells geodataframe
illegalcells_gdf = dfmt.meshkernel_get_illegalcells(mk=mk_object)
# plot
fig, ax = plt.subplots()
mk_object.mesh2d_get().plot_edges(ax,zorder=1)
# ctx.add_basemap(ax=ax, crs=crs, attribution=False)
dfmt.plot_coastlines(ax=ax, crs=crs)
>> reading coastlines: 1.61 sec
>> reading coastlines: 2.32 sec
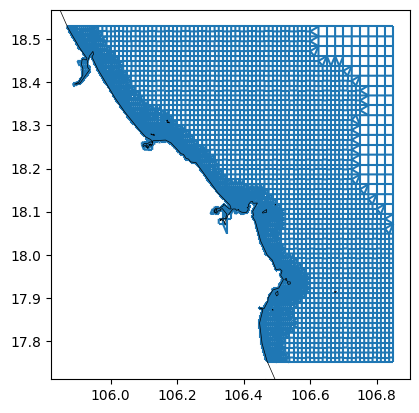
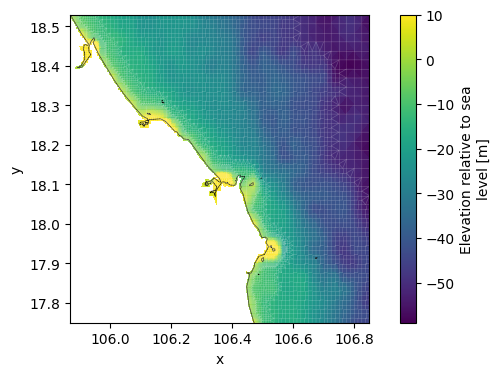
This step saves the network including bathymetry as a *_net.nc file.
[8]:
# convert to xugrid
xu_grid_uds = dfmt.meshkernel_to_UgridDataset(mk=mk_object, crs=crs)
# interpolate bathymetry onto the grid
data_bathy_interp = data_bathy_sel.interp(lon=xu_grid_uds.obj.mesh2d_node_x, lat=xu_grid_uds.obj.mesh2d_node_y)
xu_grid_uds['mesh2d_node_z'] = data_bathy_interp.clip(max=10)
# avoid _FillValue=nan: https://github.com/Deltares/dfm_tools/issues/1124
# obj needs to be accessed for this to work until fixing: https://github.com/Deltares/xugrid/issues/321
xu_grid_uds.obj.mesh2d_node_z.encoding["_FillValue"] = 1e20
# plot bathymetry and grid
fig, ax = plt.subplots(figsize=(8,4))
xu_grid_uds.mesh2d_node_z.ugrid.plot(ax=ax,center=False)
xu_grid_uds.grid.plot(ax=ax,linewidth=0.5,color='white',alpha=0.2)
# ctx.add_basemap(ax=ax, crs=crs, attribution=False)
dfmt.plot_coastlines(ax=ax, crs=crs)
# write xugrid grid to netcdf
netfile = os.path.join(dir_output, f'{model_name}_net.nc')
xu_grid_uds.ugrid.to_netcdf(netfile)
>> reading coastlines: 1.96 sec
4. Generate boundary conditions#
To simulate something useful we need boundary conditions for the model. These boundary conditions are forced via the new *.ext file. We initialize a HYDROLIB-core dflowfm.ExtModel instance and append boundaries to it.
We start with the interpolation of tidal components from a global database. In this example we use TPXO8, but other sources are also available. If you are working outside of the Deltares network, only the *_opendap sources will be available. The components of the chosen tidal model are interpolated to the points of the dflowfm.PolyFile we generated in the previous step and converted to a HYDROLIB-core dflowfm.ForcingModel (*.bc file).
The resulting tidal boundary condition has a set of tidal components with amplitudes and phases for each boundary point.
Besides tide or waterlevels, we can also add boundary conditions for flow velocities, salinity, temperature and water quality variables like NO3. In this example we download the data from the Copernicus Marine Service and interpolate it to the points of the dflowfm.PolyFile. The resulting netcdf files are also converted to a dflowfm.ForcingModel (*.bc file). The resulting boundary conditions contain a timeseries with depth dimension for
each boundary point.
Exercises#
try a different tide model
add additional WAQ variables like NO3
Boundary conditions can be supplied to the model via the new and old *.ext file. We initialize HYDROLIB-core dflowfm.ExtModel and dflowfm.ExtOldModel instance and append boundaries to it. These boundaries are HYDROLIB-core dflowfm.ExtForcing and dflowfm.ExtOldForcing instances.
[9]:
# generate new format external forcings file (*.ext): initial and open boundary condition
ext_file_new = os.path.join(dir_output, f'{model_name}_new.ext')
ext_new = hcdfm.ExtModel()
# generate old format external forcings file (*.ext): spatial data (will be moved to new ext file in 2025)
ext_file_old = os.path.join(dir_output, f'{model_name}_old.ext')
ext_old = hcdfm.ExtOldModel()
A constant waterlevel offset can be used to correct for instance for sea level rise. This results in a *.bc file with a constant value for all polylines in the plifile.
[10]:
ext_new = dfmt.constant_to_bc(ext_new=ext_new, file_pli=poly_file, constant=0.00)
Several tide models are available consisting of amplitudes and phases for several tidal components with global coverage. The ‘opendap’ sources are available for everyone, the others only within the Deltares network. This step generates a boundary conditions file (*.bc) containing amplitudes and phases for point along the model boundary.
[11]:
# interpolate tidal components to boundary conditions file (.bc)
tidemodel = 'tpxo80_opendap' # tidemodel: FES2014, FES2012, EOT20, GTSMv4.1, GTSMv4.1_opendap, tpxo80_opendap
dfmt.interpolate_tide_to_bc(ext_new=ext_new, tidemodel=tidemodel, file_pli=poly_file, component_list=None)
> interp mfdataset to all PolyFile points (lat/lon coordinates)
> actual extraction of data from netcdf with .load() (for 71 plipoints at once, this might take a while)
>>time passed: 0.00 sec
Converting 71 plipoints to hcdfm.ForcingModel(): 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71. >> done in 0.20 sec
Download spatial fields of sea surface height, salinity, temperature and currents. Convert them to boundary conditions files (*.bc) and add these to the ExtModel() object.
Exercises#
add WAQ variables like ‘no3’ (‘tracerbndNO3’) and ‘phyc’ (‘tracerbndPON1’ and others). Check
dfmt.get_conversion_dict()for an overview of parameter/quantity names.
[ ]:
# download CMEMS data
dir_output_data_cmems = os.path.join(dir_output_data, 'cmems')
os.makedirs(dir_output_data_cmems, exist_ok=True)
for varkey in ['zos','so','thetao','uo','vo']:
dfmt.download_CMEMS(varkey=varkey,
longitude_min=lon_min, longitude_max=lon_max, latitude_min=lat_min, latitude_max=lat_max,
date_min=date_min, date_max=date_max,
dir_output=dir_output_data_cmems, file_prefix='cmems_', overwrite=overwrite)
# convert to bc and add to ext
# note that if you apply 3D forcing in a 2D model, the first layer of the forcing is applied to the entire water column
list_quantities = ['waterlevelbnd','salinitybnd','temperaturebnd','uxuyadvectionvelocitybnd']
dir_pattern = os.path.join(dir_output_data_cmems,'cmems_{ncvarname}_*.nc')
ext_new = dfmt.cmems_nc_to_bc(ext_new=ext_new,
refdate_str=f'minutes since {ref_date} 00:00:00 +00:00',
dir_output=dir_output,
list_quantities=list_quantities,
tstart=date_min,
tstop=date_max,
file_pli=poly_file,
dir_pattern=dir_pattern)
INFO - 2025-06-05T09:17:36Z - Checking if credentials are valid.
INFO - 2025-06-05T09:17:37Z - Valid credentials from configuration file.
retrieving time range of CMEMS reanalysis, reanalysis-interim and forecast products (phy)
INFO - 2025-06-05T09:17:40Z - Selected dataset version: "202311"
INFO - 2025-06-05T09:17:40Z - Selected dataset part: "default"
corrected daily averaged dataset from midnight to noon by adding a 12-hour offset.
INFO - 2025-06-05T09:17:45Z - Selected dataset version: "202311"
INFO - 2025-06-05T09:17:45Z - Selected dataset part: "default"
corrected daily averaged dataset from midnight to noon by adding a 12-hour offset.
INFO - 2025-06-05T09:17:48Z - Selected dataset version: "202406"
INFO - 2025-06-05T09:17:48Z - Selected dataset part: "default"
corrected daily averaged dataset from midnight to noon by adding a 12-hour offset.
The CMEMS 'reanalysis-interim' product will be used.
downloading zos from cmems_mod_glo_phy_myint_0.083deg_P1D-m with freq=D
INFO - 2025-06-05T09:17:51Z - Selected dataset version: "202311"
INFO - 2025-06-05T09:17:51Z - Selected dataset part: "default"
INFO - 2025-06-05T09:17:53Z - Checking if credentials are valid.
corrected daily averaged dataset from midnight to noon by adding a 12-hour offset.
"cmems_zos_2022-10-31.nc" found and overwrite=False, continuing.
"cmems_zos_2022-11-01.nc" found and overwrite=False, continuing.
"cmems_zos_2022-11-02.nc" found and overwrite=False, continuing.
"cmems_zos_2022-11-03.nc" found and overwrite=False, continuing.
INFO - 2025-06-05T09:17:54Z - Valid credentials from configuration file.
The CMEMS 'reanalysis-interim' product will be used.
downloading so from cmems_mod_glo_phy_myint_0.083deg_P1D-m with freq=D
INFO - 2025-06-05T09:17:54Z - Selected dataset version: "202311"
INFO - 2025-06-05T09:17:54Z - Selected dataset part: "default"
INFO - 2025-06-05T09:17:56Z - Checking if credentials are valid.
corrected daily averaged dataset from midnight to noon by adding a 12-hour offset.
"cmems_so_2022-10-31.nc" found and overwrite=False, continuing.
"cmems_so_2022-11-01.nc" found and overwrite=False, continuing.
"cmems_so_2022-11-02.nc" found and overwrite=False, continuing.
"cmems_so_2022-11-03.nc" found and overwrite=False, continuing.
INFO - 2025-06-05T09:17:57Z - Valid credentials from configuration file.
The CMEMS 'reanalysis-interim' product will be used.
downloading thetao from cmems_mod_glo_phy_myint_0.083deg_P1D-m with freq=D
INFO - 2025-06-05T09:17:58Z - Selected dataset version: "202311"
INFO - 2025-06-05T09:17:58Z - Selected dataset part: "default"
INFO - 2025-06-05T09:18:00Z - Checking if credentials are valid.
corrected daily averaged dataset from midnight to noon by adding a 12-hour offset.
"cmems_thetao_2022-10-31.nc" found and overwrite=False, continuing.
"cmems_thetao_2022-11-01.nc" found and overwrite=False, continuing.
"cmems_thetao_2022-11-02.nc" found and overwrite=False, continuing.
"cmems_thetao_2022-11-03.nc" found and overwrite=False, continuing.
INFO - 2025-06-05T09:18:01Z - Valid credentials from configuration file.
The CMEMS 'reanalysis-interim' product will be used.
downloading uo from cmems_mod_glo_phy_myint_0.083deg_P1D-m with freq=D
INFO - 2025-06-05T09:18:01Z - Selected dataset version: "202311"
INFO - 2025-06-05T09:18:01Z - Selected dataset part: "default"
INFO - 2025-06-05T09:18:03Z - Checking if credentials are valid.
corrected daily averaged dataset from midnight to noon by adding a 12-hour offset.
"cmems_uo_2022-10-31.nc" found and overwrite=False, continuing.
"cmems_uo_2022-11-01.nc" found and overwrite=False, continuing.
"cmems_uo_2022-11-02.nc" found and overwrite=False, continuing.
"cmems_uo_2022-11-03.nc" found and overwrite=False, continuing.
INFO - 2025-06-05T09:18:05Z - Valid credentials from configuration file.
The CMEMS 'reanalysis-interim' product will be used.
downloading vo from cmems_mod_glo_phy_myint_0.083deg_P1D-m with freq=D
INFO - 2025-06-05T09:18:05Z - Selected dataset version: "202311"
INFO - 2025-06-05T09:18:05Z - Selected dataset part: "default"
corrected daily averaged dataset from midnight to noon by adding a 12-hour offset.
"cmems_vo_2022-10-31.nc" found and overwrite=False, continuing.
"cmems_vo_2022-11-01.nc" found and overwrite=False, continuing.
"cmems_vo_2022-11-02.nc" found and overwrite=False, continuing.
"cmems_vo_2022-11-03.nc" found and overwrite=False, continuing.
processing quantity: waterlevelbnd
loading mfdataset of 4 files with pattern(s) c:\DATA\checkouts\dfm_tools\docs\notebooks\Vietnam_2D_model\data\cmems\cmems_zos_*.nc
variable zos renamed to waterlevelbnd
> interp mfdataset to all PolyFile points (lat/lon coordinates)
> actual extraction of data from netcdf with .load() (for 71 plipoints at once, this might take a while)
>>time passed: 0.04 sec
Converting 71 plipoints to hcdfm.ForcingModel(): 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71. >> done in 0.06 sec
processing quantity: salinitybnd
loading mfdataset of 4 files with pattern(s) c:\DATA\checkouts\dfm_tools\docs\notebooks\Vietnam_2D_model\data\cmems\cmems_so_*.nc
dimension depth renamed to z
varname depth renamed to z
variable so renamed to salinitybnd
> interp mfdataset to all PolyFile points (lat/lon coordinates)
> actual extraction of data from netcdf with .load() (for 71 plipoints at once, this might take a while)
>>time passed: 0.03 sec
Converting 71 plipoints to hcdfm.ForcingModel(): 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71. >> done in 0.14 sec
processing quantity: temperaturebnd
loading mfdataset of 4 files with pattern(s) c:\DATA\checkouts\dfm_tools\docs\notebooks\Vietnam_2D_model\data\cmems\cmems_thetao_*.nc
dimension depth renamed to z
varname depth renamed to z
variable thetao renamed to temperaturebnd
> interp mfdataset to all PolyFile points (lat/lon coordinates)
> actual extraction of data from netcdf with .load() (for 71 plipoints at once, this might take a while)
>>time passed: 0.03 sec
Converting 71 plipoints to hcdfm.ForcingModel(): 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71. >> done in 0.20 sec
processing quantity: uxuyadvectionvelocitybnd
loading mfdataset of 4 files with pattern(s) c:\DATA\checkouts\dfm_tools\docs\notebooks\Vietnam_2D_model\data\cmems\cmems_uo_*.nc
dimension depth renamed to z
varname depth renamed to z
variable uo renamed to ux
loading mfdataset of 4 files with pattern(s) c:\DATA\checkouts\dfm_tools\docs\notebooks\Vietnam_2D_model\data\cmems\cmems_vo_*.nc
dimension depth renamed to z
varname depth renamed to z
variable vo renamed to uy
> interp mfdataset to all PolyFile points (lat/lon coordinates)
> actual extraction of data from netcdf with .load() (for 71 plipoints at once, this might take a while)
>>time passed: 0.06 sec
57 58 59 60 61 62 63 64 65 66 67 68 69 70 71. >> done in 0.39 sec 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56
[13]:
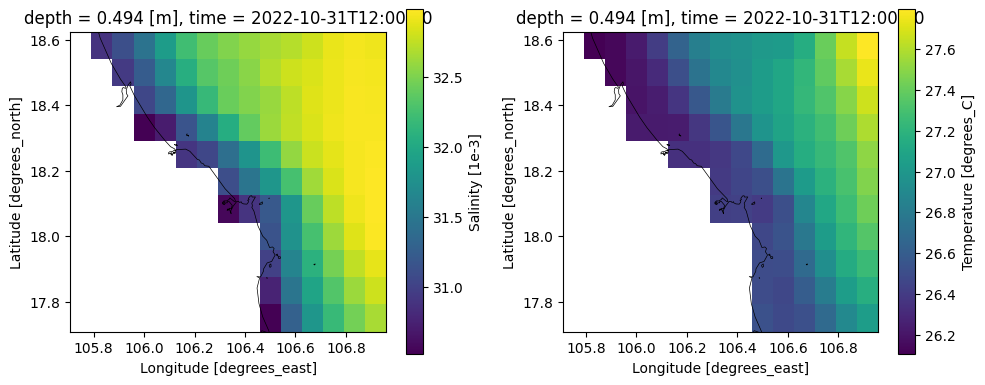
# plot downloaded CMEMS data
file_cmems = os.path.join(dir_output_data,'cmems','*.nc')
ds_cmems = xr.open_mfdataset(file_cmems)
ds_cmems
# plot
fig, (ax1,ax2) = plt.subplots(1, 2, figsize=(10,4))
ds_cmems.so.isel(time=0, depth=0).plot(ax=ax1)
dfmt.plot_coastlines(ax=ax1, crs=crs)
ds_cmems.thetao.isel(time=0, depth=0).plot(ax=ax2)
dfmt.plot_coastlines(ax=ax2, crs=crs)
fig.tight_layout()
>> reading coastlines: 1.82 sec
2.04 secng coastlines:
[14]:
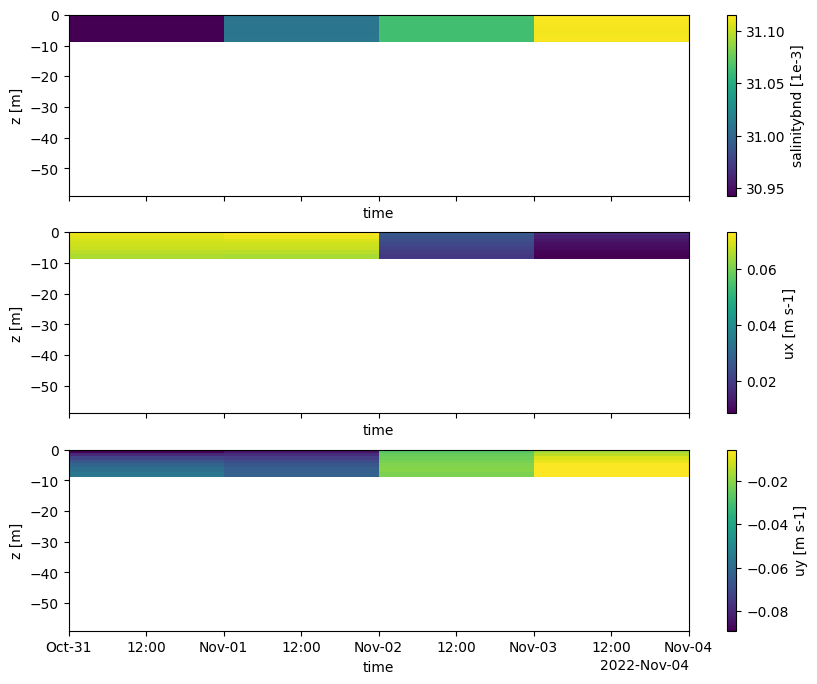
# plot interpolated CMEMS data (boundary conditions in .bc)
pli_basename = Path(poly_file).stem
file_bc_sal = os.path.join(dir_output, f'salinitybnd_CMEMS_{pli_basename}.bc')
bc_obj_sal = hcdfm.ForcingModel(file_bc_sal)
forcing_xr_sal = dfmt.forcinglike_to_Dataset(bc_obj_sal.forcing[0], convertnan=True)
file_bc_uxuy = os.path.join(dir_output,f'uxuyadvectionvelocitybnd_CMEMS_{pli_basename}.bc')
bc_obj_uxuy = hcdfm.ForcingModel(file_bc_uxuy)
forcing_xr_uxuy = dfmt.forcinglike_to_Dataset(bc_obj_uxuy.forcing[0], convertnan=True)
# plot
fig, (ax1,ax2,ax3) = plt.subplots(3, 1, sharex=True, sharey=True, figsize=(10,8))
forcing_xr_sal['salinitybnd'].T.plot(ax=ax1)
forcing_xr_uxuy['ux'].T.plot(ax=ax2)
forcing_xr_uxuy['uy'].T.plot(ax=ax3)
ax1.set_ylim(xu_grid_uds.mesh2d_node_z.min(), None)
[14]:
(-58.96910790071473, 0.02965003252029419)
In order to decrease the necessary model spinup time we can use spatially varying initial conditions. We derive these again from the data downloaded from the Copernicus Marine Service. The netcdf files are subsetted in time and written to the netcdf format expected by D-Flow FM. This step writes *.nc files containing initial fields.
Initial fields for salinity and temperature can only be added in case of 3D model and mdu.physics.iniwithnudge=2. Via this same function it is possible to add initial conditions for tracers, but not yet for the waterlevel and velocity field.
Excercises#
add initial condtions for WAQ parameters like ‘tracerbndNO3’ and ‘tracerbndPON1’
[15]:
# convert downloaded CMEMS data to initial fields
if modeltype == "3D":
ext_old = dfmt.cmems_nc_to_ini(ext_old=ext_old,
dir_output=dir_output,
list_quantities=['salinitybnd','temperaturebnd'], # list_quantities,
tstart=date_min,
dir_pattern=dir_pattern)
Another important forcing is meteo. In this example ERA5 meteo data is retrieved from the Copernicus Climate Data Store. The netcdf files are merged in time and written to the netcdf format expected by D-Flow FM. This step generates *.nc files containing the meteo data.
Exercises#
in case of a 3D model with salinity and temperature enabled, you could add more meteo parameters. In that case also enable the composit ocean model via
mdu.physics.temperature = 5.
[16]:
# ERA5 - download spatial fields of air pressure, wind speeds and Charnock coefficient
dir_output_data_era5 = os.path.join(dir_output_data, 'ERA5')
os.makedirs(dir_output_data_era5, exist_ok=True)
varkey_list = ['msl','u10n','v10n','chnk']
# for 3D models a more extensive forcing can be considered
# this includes dewpoint, airtemperature, cloudiness, solarradiation, longwaveradiation, evaporation, precipitation
# if modeltype == "3D":
# varkey_list = ['msl','u10n','v10n','chnk','d2m','t2m','tcc','ssr','strd','mer','mtpr']
for varkey in varkey_list:
dfmt.download_ERA5(varkey,
longitude_min=lon_min, longitude_max=lon_max, latitude_min=lat_min, latitude_max=lat_max,
date_min=date_min, date_max=date_max,
dir_output=dir_output_data_era5, overwrite=overwrite)
# ERA5 meteo - convert to netCDF for usage in Delft3D FM
ext_old = dfmt.preprocess_merge_meteofiles_era5(ext_old=ext_old,
varkey_list=varkey_list,
dir_data=dir_output_data_era5,
dir_output=dir_output,
time_slice=slice(date_min, date_max))
2025-06-05 09:18:17,246 INFO [2024-09-26T00:00:00] Watch our [Forum](https://forum.ecmwf.int/) for Announcements, news and other discussed topics.
2025-06-05 09:18:17,436 INFO [2024-09-26T00:00:00] Watch our [Forum](https://forum.ecmwf.int/) for Announcements, news and other discussed topics.
found ECMWF API-key and authorization successful
retrieving data from 2022-11 to 2022-11 (freq=<MonthEnd>)
"era5_msl_2022-11.nc" found and overwrite=False, continuing.
2025-06-05 09:18:17,557 INFO [2024-09-26T00:00:00] Watch our [Forum](https://forum.ecmwf.int/) for Announcements, news and other discussed topics.
2025-06-05 09:18:17,743 INFO [2024-09-26T00:00:00] Watch our [Forum](https://forum.ecmwf.int/) for Announcements, news and other discussed topics.
found ECMWF API-key and authorization successful
retrieving data from 2022-11 to 2022-11 (freq=<MonthEnd>)
"era5_u10n_2022-11.nc" found and overwrite=False, continuing.
2025-06-05 09:18:17,874 INFO [2024-09-26T00:00:00] Watch our [Forum](https://forum.ecmwf.int/) for Announcements, news and other discussed topics.
2025-06-05 09:18:18,067 INFO [2024-09-26T00:00:00] Watch our [Forum](https://forum.ecmwf.int/) for Announcements, news and other discussed topics.
found ECMWF API-key and authorization successful
retrieving data from 2022-11 to 2022-11 (freq=<MonthEnd>)
"era5_v10n_2022-11.nc" found and overwrite=False, continuing.
2025-06-05 09:18:18,196 INFO [2024-09-26T00:00:00] Watch our [Forum](https://forum.ecmwf.int/) for Announcements, news and other discussed topics.
2025-06-05 09:18:18,375 INFO [2024-09-26T00:00:00] Watch our [Forum](https://forum.ecmwf.int/) for Announcements, news and other discussed topics.
found ECMWF API-key and authorization successful
retrieving data from 2022-11 to 2022-11 (freq=<MonthEnd>)
"era5_chnk_2022-11.nc" found and overwrite=False, continuing.
>> opening multifile dataset of 1 files (can take a while with lots of files): 0.02 sec
>> writing file (can take a while): 0.02 sec
>> opening multifile dataset of 1 files (can take a while with lots of files): 0.01 sec
0.02 secng file (can take a while):
>> opening multifile dataset of 1 files (can take a while with lots of files): 0.01 sec
>> writing file (can take a while): 0.02 sec
>> opening multifile dataset of 1 files (can take a while with lots of files): 0.04 sec
>> writing file (can take a while): 0.02 sec
[17]:
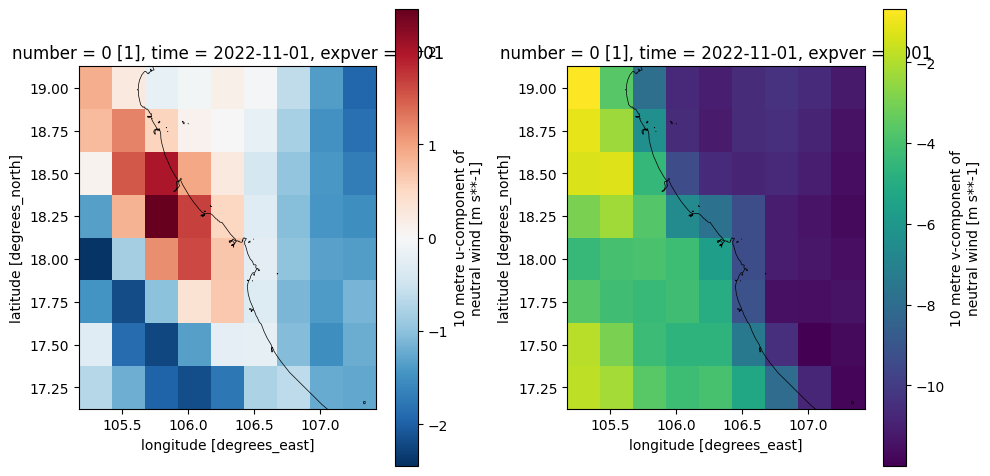
# plot converted ERA5 data
file_era5 = os.path.join(dir_output,'era5*.nc')
ds_era5 = xr.open_mfdataset(file_era5)
ds_era5
# plot
fig, (ax1,ax2) = plt.subplots(1,2,figsize=(10,5))
ds_era5.u10n.isel(time=0).plot(ax=ax1)
dfmt.plot_coastlines(ax=ax1, crs=crs)
ds_era5.v10n.isel(time=0).plot(ax=ax2)
dfmt.plot_coastlines(ax=ax2, crs=crs)
fig.tight_layout()
>> reading coastlines: 2.53 sec
>> reading coastlines: 3.57 sec
These are saved as .ext files. They contain all the paths to the model forcing and have to be linked in the .mdu file below.
[18]:
# save new ext file
ext_new.save(filepath=ext_file_new) # ,path_style=path_style)
# save old ext file
ext_old.save(filepath=ext_file_old) # , path_style=path_style)
6. Generate obsfile#
The D-Flow FM model wil have mapoutput and hisoutput. In order for the hisoutput to be produced, we need to specify the locations where output will be saved. These locations need to be available via a *_obs.xyn file containing all observation points in the model domain.
We base the default selection of the observation points on the set of locations used in the GTSM-ERA5 dataset (Global Tide and Surge Model forced with ERA5 reanalysis). This dataset contains over 43000 points along global coastlines and (gridded) in open waters.
Exercises#
the observation points are by default based on the observation points GTSM-ERA5 dataset, try adding also the obspoints near specific cities/harbours/islands (variable
obs_pd)
[19]:
# Subset observation points used in the GTSM-ERA5 dataset
gtsm_locs_gpd = dfmt.ssh_catalog_subset(source='gtsm3-era5-cds', lon_min=lon_min, lon_max=lon_max, lat_min=lat_min, lat_max=lat_max)
# Convert the dataframe to the file format that can be read by D-Flow FM and save file
file_obs = os.path.join(dir_output, f'{model_name}_obs.xyn')
dfmt.ssh_catalog_toxynfile(gtsm_locs_gpd, file_obs)
# Load the .xyn file with the observation points (created above) to view contents
obs_pd = pd.read_csv(file_obs, sep=r'\s+', header=None, names=['x','y','name'])
obs_pd
[19]:
| x | y | name | |
|---|---|---|---|
| 0 | 105.923 | 18.472 | gtsm3-era5-16615-id_coast_glob_16662 |
| 1 | 106.069 | 18.325 | gtsm3-era5-16616-id_coast_glob_16663 |
| 2 | 106.245 | 18.208 | gtsm3-era5-16617-id_coast_glob_16664 |
| 3 | 106.450 | 18.091 | gtsm3-era5-16618-id_coast_glob_16665 |
| 4 | 106.479 | 17.915 | gtsm3-era5-16619-id_coast_glob_16666 |
| 5 | 106.128 | 18.267 | gtsm3-era5-34915-id_reg_grid_glob_01959 |
| 6 | 106.333 | 18.120 | gtsm3-era5-40529-id_obs_vung_ang_651a_vietnam_... |
[20]:
# Visualize the locations of the points on the model grid
fig, ax = plt.subplots(figsize=(8,4))
xu_grid_uds.grid.plot(ax=ax, linewidth=0.5, color='lightgray')
ax.plot(obs_pd['x'], obs_pd['y'], 'rx')
dfmt.plot_coastlines(ax=ax, crs=crs)
>> reading coastlines: 2.31 sec
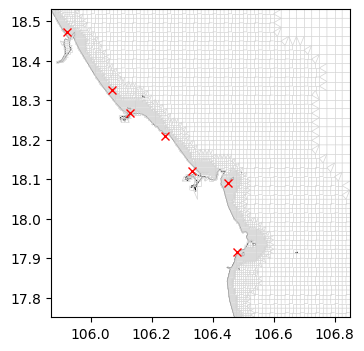
7. Generate mdu file#
In order for the model to run, we need a model definition file. In case of D-Flow FM this is a .mdu file, represented by the HYDROLIB-core ``dflowfm.FMModel``. This is initialized and many of the files we generated above (network, extfiles, obsfile) are linked here. You can visualize the resulting model tree with ``mdu.show_tree()``. We can also adjust any of the model parameters by setting them below. At the end of this code block the model settings are saved in the .mdu file.
Exercises#
convert this 2D model into a 3D model with the
modeltypevariable all the way aboveenable the computation of salinity and temperature in the mdu (
salinity=1 andtemperature=1)enable the composite ocean model (
temperature=5), this requires additional meteo forcing to be activated aboveuse the fourier smoothing time (
tlfSmo) to make the model spinup less chaotic
[21]:
# initialize mdu file and update settings
# all keywords are documented in the Delft3D FM user manual
# additional 2D/3D settings discussed in https://github.com/Deltares/dfm_tools/issues/951
mdu_file = os.path.join(dir_output, f'{model_name}.mdu')
mdu = hcdfm.FMModel()
# add the grid (_net.nc, network file)
mdu.geometry.netfile = netfile
mdu.geometry.openboundarytolerance = 0.1
mdu.geometry.bedlevuni = 5 # default of -5 may cause instabilities at coastline
mdu.geometry.dxwuimin2d = 0.1 # improved stability in triangular network cells
# create and add drypointsfile if there are any cells generated that will result in high orthogonality
if len(illegalcells_gdf) > 0:
illegalcells_polyfile = dfmt.geodataframe_to_PolyFile(illegalcells_gdf)
illegalcells_file = os.path.join(dir_output, "illegalcells.pol")
illegalcells_polyfile.save(illegalcells_file)
mdu.geometry.drypointsfile = [illegalcells_polyfile]
# update numerics settings
mdu.numerics.izbndpos = 1 # boundary points are on network boundary
mdu.numerics.mintimestepbreak = 0.1 # causes instable model to crash
mdu.numerics.keepstbndonoutflow = 1 # s/t on outflow boundaries, should be new default
mdu.numerics.barocponbnd = 1 # enable baroclinic pressure gradient on open boundaries, should be new default
mdu.numerics.vertadvtypsal = 4 # should be new default (is currently 6)
mdu.numerics.vertadvtyptem = 4 # should be new default (is currently 6)
# update physics settings
mdu.physics.rhomean = 1023. # for coastal models
# update wind settings
mdu.wind.icdtyp = 4 # Charnock in case of ERA5
mdu.wind.cdbreakpoints = [0.025] # important if icdtyp=4, but 0.018 or 0.041 might be better. Value is overwritten by spacevarying charnock from ERA5.
mdu.wind.pavbnd = 101330 # for inverse barometer correction, important in case of CMEMS boundary conditions
# update time settings
mdu.time.refdate = pd.Timestamp(ref_date).strftime('%Y%m%d')
mdu.time.tunit = 'S'
mdu.time.dtmax = 30
mdu.time.startdatetime = pd.Timestamp(date_min).strftime('%Y%m%d%H%M%S')
mdu.time.stopdatetime = pd.Timestamp(date_max).strftime('%Y%m%d%H%M%S')
# add the external forcing files (.ext)
mdu.external_forcing.extforcefile = ext_file_old
# use extobject instead of extpath until fix of https://github.com/Deltares/HYDROLIB-core/issues/516
mdu.external_forcing.extforcefilenew = ext_new
# update output settings (many additional settings can decrease file size significantly)
mdu.output.obsfile = [file_obs]
mdu.output.hisinterval = [600] # 10 minutes
mdu.output.mapinterval = [1800] # 30 minutes
mdu.output.rstinterval = [0] # disable restart netcdf output
mdu.output.statsinterval = [3600] # regular progress statistics in the dia file
if modeltype == "3D":
mdu.geometry.layertype = 1 # sigma-layers (default value)
mdu.geometry.kmx = 10 # number of layers
mdu.physics.iniwithnudge = 2 # support for initial sal/tem fields via iniwithnudge (requires a 3D model)
mdu.physics.salinity = 1 # enable salinity
mdu.physics.temperature = 5 # composite ocean model. This is automatically set to 1 if additional ERA5 variables are not provided.
mdu.physics.initialsalinity = 30 # for coastal models (default is 0)
mdu.physics.heat_eachstep = 1 # compute heatflux model on each timestep
mdu.wind.rhowaterinwindstress = 1 # better wind/water interaction, was called mdu.physics.rhoairrhowater before Delft3D FM 2026.01
# save .mdu file
mdu.save(mdu_file) # ,path_style=path_style)
# make all paths relative (might be properly implemented in https://github.com/Deltares/HYDROLIB-core/issues/532)
# currently only works with path_style='windows' (or actually: same OS as IDE)
dfmt.make_paths_relative(mdu_file)
8. Generate DIMR and bat file#
In order to run the model via DIMR we need a dimr_config.xml file. If you are running this notebook on a Windows platform, a .bat file will also be created with which you can run the model directly. In order for this to work you need to update the ``dimrset_folder`` to the path where the ``x64`` and or ``lnx64`` folder is located. Provide ``None`` if you have no D-Flow FM executable available on your system. This step generates a dimr_config.xml file and optionally also a .bat file.
[22]:
nproc = 1 # number of processes
dimrset_folder = None # alternatively r"c:\Program Files\Deltares\Delft3D FM Suite 2026.01 HMWQ\plugins\DeltaShell.Dimr\kernels" #alternatively r"p:\d-hydro\dimrset\weekly\2.29.28_delft3dfm_2026_01"
dfmt.create_model_exec_files(file_mdu=mdu_file, nproc=nproc, dimrset_folder=dimrset_folder)
writing dimr_config.xml
no dimrset_folder provided, cannot write bat/sh file
9. Visualize model tree#
[23]:
# visualize the model tree, show_tree is available for all HYDROLIB-core model components
mdu_obj = hcdfm.FMModel(mdu_file)
mdu_obj.show_tree()
c:\DATA\checkouts\dfm_tools\docs\notebooks\Vietnam_2D_model\Vietnam.mdu
Geometry
∟ Vietnam_net.nc
ExternalForcing
∟ Vietnam_old.ext
∟ ExtOldForcing
∟ era5_msl_20221101to20221103_ERA5.nc
∟ ExtOldForcing
∟ era5_u10n_20221101to20221103_ERA5.nc
∟ ExtOldForcing
∟ era5_v10n_20221101to20221103_ERA5.nc
∟ ExtOldForcing
∟ era5_chnk_20221101to20221103_ERA5.nc
∟ Vietnam_new.ext
∟ Boundary
∟ Vietnam.pli
∟ waterlevelbnd_constant_Vietnam.bc
∟ Boundary
∟ Vietnam.pli
∟ tide_tpxo80_opendap_Vietnam.bc
∟ Boundary
∟ Vietnam.pli
∟ waterlevelbnd_CMEMS_Vietnam.bc
∟ Boundary
∟ Vietnam.pli
∟ salinitybnd_CMEMS_Vietnam.bc
∟ Boundary
∟ Vietnam.pli
∟ temperaturebnd_CMEMS_Vietnam.bc
∟ Boundary
∟ Vietnam.pli
∟ uxuyadvectionvelocitybnd_CMEMS_Vietnam.bc
Output
∟ Vietnam_obs.xyn
10. Run the model and do the exercises#
Running the model and post-processing the results#
run the model with the
run_parallel.batfile if you work on Windows.the modeloutput can visualized with (the code in) the postprocessing notebook.
Exercises#
you can change the model input as suggested in the exercises above and rerun the model builder and the model.
read the D-Flow FM manual for background information.
Advanced exercises#
add a observation cross-section near a harbour or river
add a river inflow as a boundary condition (.pli and .bc in new format .ext or .pli and .tim in old format .ext)