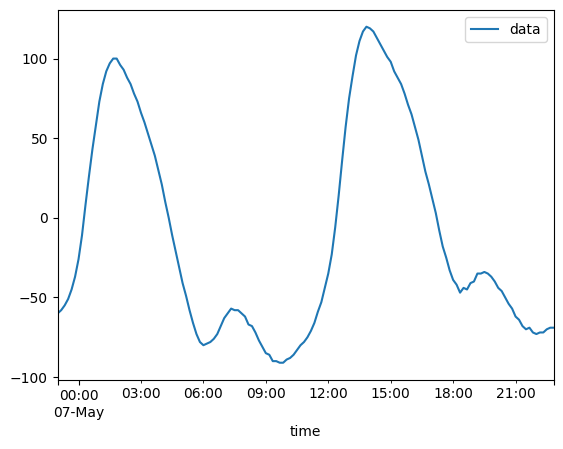
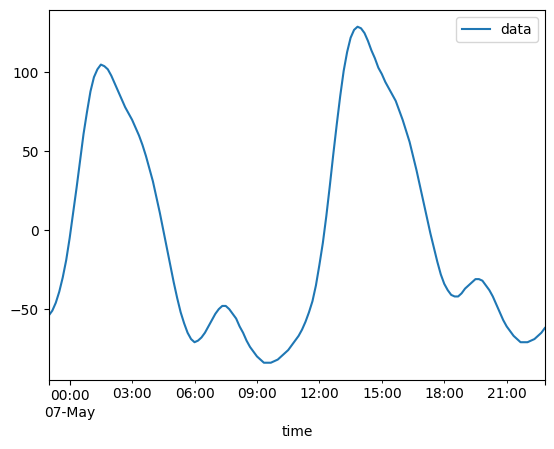
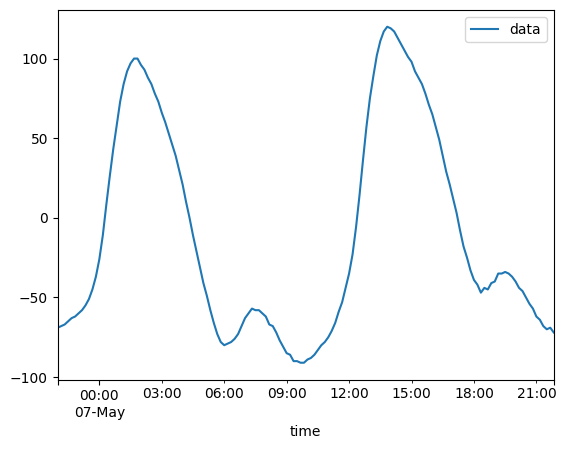
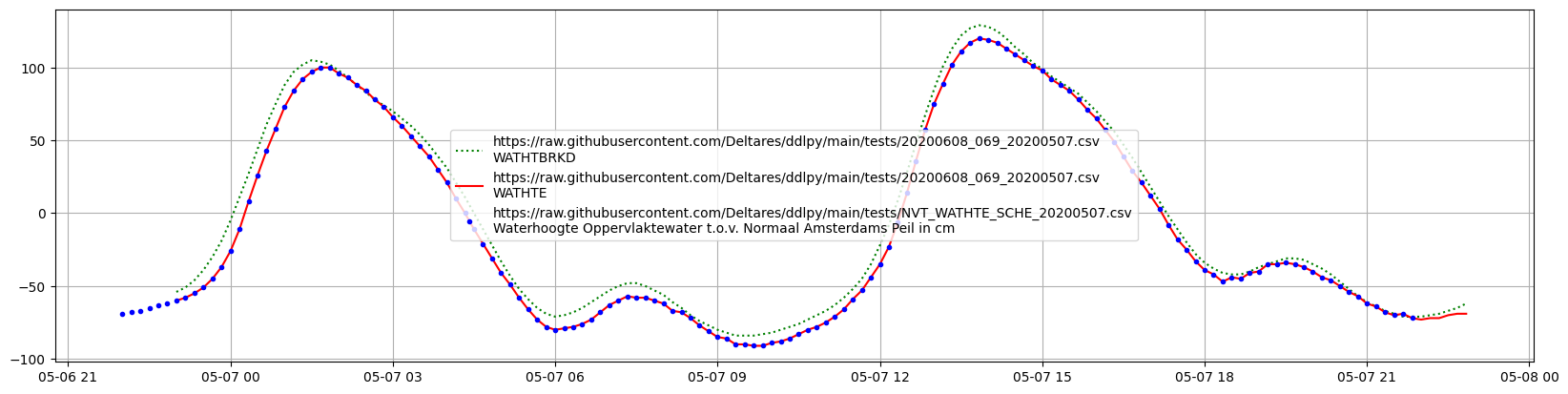
2020-05-06T23:00:00 ... 2020-05-...
array(['2020-05-06T23:00:00.000000000', '2020-05-06T23:10:00.000000000',
'2020-05-06T23:20:00.000000000', '2020-05-06T23:30:00.000000000',
'2020-05-06T23:40:00.000000000', '2020-05-06T23:50:00.000000000',
'2020-05-07T00:00:00.000000000', '2020-05-07T00:10:00.000000000',
'2020-05-07T00:20:00.000000000', '2020-05-07T00:30:00.000000000',
'2020-05-07T00:40:00.000000000', '2020-05-07T00:50:00.000000000',
'2020-05-07T01:00:00.000000000', '2020-05-07T01:10:00.000000000',
'2020-05-07T01:20:00.000000000', '2020-05-07T01:30:00.000000000',
'2020-05-07T01:40:00.000000000', '2020-05-07T01:50:00.000000000',
'2020-05-07T02:00:00.000000000', '2020-05-07T02:10:00.000000000',
'2020-05-07T02:20:00.000000000', '2020-05-07T02:30:00.000000000',
'2020-05-07T02:40:00.000000000', '2020-05-07T02:50:00.000000000',
'2020-05-07T03:00:00.000000000', '2020-05-07T03:10:00.000000000',
'2020-05-07T03:20:00.000000000', '2020-05-07T03:30:00.000000000',
'2020-05-07T03:40:00.000000000', '2020-05-07T03:50:00.000000000',
'2020-05-07T04:00:00.000000000', '2020-05-07T04:10:00.000000000',
'2020-05-07T04:20:00.000000000', '2020-05-07T04:30:00.000000000',
'2020-05-07T04:40:00.000000000', '2020-05-07T04:50:00.000000000',
'2020-05-07T05:00:00.000000000', '2020-05-07T05:10:00.000000000',
'2020-05-07T05:20:00.000000000', '2020-05-07T05:30:00.000000000',
'2020-05-07T05:40:00.000000000', '2020-05-07T05:50:00.000000000',
'2020-05-07T06:00:00.000000000', '2020-05-07T06:10:00.000000000',
'2020-05-07T06:20:00.000000000', '2020-05-07T06:30:00.000000000',
'2020-05-07T06:40:00.000000000', '2020-05-07T06:50:00.000000000',
'2020-05-07T07:00:00.000000000', '2020-05-07T07:10:00.000000000',
'2020-05-07T07:20:00.000000000', '2020-05-07T07:30:00.000000000',
'2020-05-07T07:40:00.000000000', '2020-05-07T07:50:00.000000000',
'2020-05-07T08:00:00.000000000', '2020-05-07T08:10:00.000000000',
'2020-05-07T08:20:00.000000000', '2020-05-07T08:30:00.000000000',
'2020-05-07T08:40:00.000000000', '2020-05-07T08:50:00.000000000',
'2020-05-07T09:00:00.000000000', '2020-05-07T09:10:00.000000000',
'2020-05-07T09:20:00.000000000', '2020-05-07T09:30:00.000000000',
'2020-05-07T09:40:00.000000000', '2020-05-07T09:50:00.000000000',
'2020-05-07T10:00:00.000000000', '2020-05-07T10:10:00.000000000',
'2020-05-07T10:20:00.000000000', '2020-05-07T10:30:00.000000000',
'2020-05-07T10:40:00.000000000', '2020-05-07T10:50:00.000000000',
'2020-05-07T11:00:00.000000000', '2020-05-07T11:10:00.000000000',
'2020-05-07T11:20:00.000000000', '2020-05-07T11:30:00.000000000',
'2020-05-07T11:40:00.000000000', '2020-05-07T11:50:00.000000000',
'2020-05-07T12:00:00.000000000', '2020-05-07T12:10:00.000000000',
'2020-05-07T12:20:00.000000000', '2020-05-07T12:30:00.000000000',
'2020-05-07T12:40:00.000000000', '2020-05-07T12:50:00.000000000',
'2020-05-07T13:00:00.000000000', '2020-05-07T13:10:00.000000000',
'2020-05-07T13:20:00.000000000', '2020-05-07T13:30:00.000000000',
'2020-05-07T13:40:00.000000000', '2020-05-07T13:50:00.000000000',
'2020-05-07T14:00:00.000000000', '2020-05-07T14:10:00.000000000',
'2020-05-07T14:20:00.000000000', '2020-05-07T14:30:00.000000000',
'2020-05-07T14:40:00.000000000', '2020-05-07T14:50:00.000000000',
'2020-05-07T15:00:00.000000000', '2020-05-07T15:10:00.000000000',
'2020-05-07T15:20:00.000000000', '2020-05-07T15:30:00.000000000',
'2020-05-07T15:40:00.000000000', '2020-05-07T15:50:00.000000000',
'2020-05-07T16:00:00.000000000', '2020-05-07T16:10:00.000000000',
'2020-05-07T16:20:00.000000000', '2020-05-07T16:30:00.000000000',
'2020-05-07T16:40:00.000000000', '2020-05-07T16:50:00.000000000',
'2020-05-07T17:00:00.000000000', '2020-05-07T17:10:00.000000000',
'2020-05-07T17:20:00.000000000', '2020-05-07T17:30:00.000000000',
'2020-05-07T17:40:00.000000000', '2020-05-07T17:50:00.000000000',
'2020-05-07T18:00:00.000000000', '2020-05-07T18:10:00.000000000',
'2020-05-07T18:20:00.000000000', '2020-05-07T18:30:00.000000000',
'2020-05-07T18:40:00.000000000', '2020-05-07T18:50:00.000000000',
'2020-05-07T19:00:00.000000000', '2020-05-07T19:10:00.000000000',
'2020-05-07T19:20:00.000000000', '2020-05-07T19:30:00.000000000',
'2020-05-07T19:40:00.000000000', '2020-05-07T19:50:00.000000000',
'2020-05-07T20:00:00.000000000', '2020-05-07T20:10:00.000000000',
'2020-05-07T20:20:00.000000000', '2020-05-07T20:30:00.000000000',
'2020-05-07T20:40:00.000000000', '2020-05-07T20:50:00.000000000',
'2020-05-07T21:00:00.000000000', '2020-05-07T21:10:00.000000000',
'2020-05-07T21:20:00.000000000', '2020-05-07T21:30:00.000000000',
'2020-05-07T21:40:00.000000000', '2020-05-07T21:50:00.000000000',
'2020-05-07T22:00:00.000000000', '2020-05-07T22:10:00.000000000',
'2020-05-07T22:20:00.000000000', '2020-05-07T22:30:00.000000000',
'2020-05-07T22:40:00.000000000', '2020-05-07T22:50:00.000000000'],
dtype='datetime64[ns]')