Utilities API
FM2PROF includes a set of utilities to analyse its output.
Compare1D2D
Bases: ModelOutputReader
Utility to compare the results of a 1D and 2D model through visualisation and statistical post-processing.
Note
If 2D and 1D netCDF input files are provided, they will first be
converted to csv files. Once csv files are present, the original
netCDF files are no longer used. In that case, the arguments
to path_1d and path_2d should be None.
Example usage
Parameters
project: `fm2prof.Project` object.
path_1d: path to SOBEK dimr directory
path_2d: path to his nc file
routes: list of branch abbreviations, e.g. ['NR', 'LK']
start_time: start time for plotting and analytics. Use this to crop the time to prevent initalisation from
affecting statistics.
stop_time: stop time for plotting and analytics.
style: `PlotStyles` style
Instantiate a Compare1D2D object.
Source code in fm2prof\utils.py
1592 1593 1594 1595 1596 1597 1598 1599 1600 1601 1602 1603 1604 1605 1606 1607 1608 1609 1610 1611 1612 1613 1614 1615 1616 1617 1618 1619 1620 1621 1622 1623 1624 1625 1626 1627 1628 1629 1630 1631 1632 1633 1634 1635 1636 1637 1638 1639 1640 1641 1642 1643 1644 1645 1646 1647 1648 1649 1650 1651 1652 1653 1654 1655 | |
colorscheme: str
property
Color scheme.
file_1d_h_digitized: Path
property
Return 1D water level digitized file path.
file_2d_h_digitized: Path
property
Return 2D water level digitized file path.
routes: list[list[str]]
property
writable
Return routes.
digitize_data()
Compute the average for a given bin for 1D and 2D water level data.
Use to make Q-H graphs instead of T-H graph
Source code in fm2prof\utils.py
eval()
Create multiple figures.
Source code in fm2prof\utils.py
figure_at_station(station, func='time', *, savefig=True)
Create a figure with the timeseries at a single observation station.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
station
|
str
|
name of station. use |
required |
func
|
str
|
use |
'time'
|
savefig
|
bool
|
if True, saves to png. If False, returned FigureOutput. Defaults to True. |
True
|
Returns:
| Type | Description |
|---|---|
FigureOutput | None
|
FigureOutput | None: FigureOutput object or None if savefig is set to True. |
Source code in fm2prof\utils.py
1804 1805 1806 1807 1808 1809 1810 1811 1812 1813 1814 1815 1816 1817 1818 1819 1820 1821 1822 1823 1824 1825 1826 1827 1828 1829 1830 1831 1832 1833 1834 1835 1836 1837 1838 1839 1840 1841 1842 1843 1844 1845 1846 1847 1848 1849 1850 1851 1852 1853 1854 1855 1856 1857 1858 1859 1860 1861 1862 1863 1864 1865 1866 1867 1868 1869 1870 1871 1872 1873 1874 1875 1876 1877 1878 1879 1880 1881 1882 1883 1884 1885 1886 1887 1888 1889 1890 1891 | |
figure_compare_discharge_at_stations(stations, title='no_title', *, savefig=True)
Comparea discharge distribution over two stations.
Like Compare1D2D.figure_at_station.
Example usage:
Figures are saved to[Compare1D2D.output_path]/figures/discharge
Example output:
.. figure:: figures_utils/discharge/example.png
Example output figure
Source code in fm2prof\utils.py
1945 1946 1947 1948 1949 1950 1951 1952 1953 1954 1955 1956 1957 1958 1959 1960 1961 1962 1963 1964 1965 1966 1967 1968 1969 1970 1971 1972 1973 1974 1975 1976 1977 1978 1979 1980 1981 1982 1983 1984 1985 1986 1987 1988 1989 1990 1991 1992 1993 1994 1995 1996 1997 1998 1999 2000 2001 2002 2003 2004 2005 2006 2007 2008 2009 2010 2011 2012 2013 2014 2015 2016 2017 2018 2019 2020 2021 2022 2023 2024 2025 2026 2027 2028 2029 2030 2031 2032 2033 2034 2035 2036 2037 2038 2039 2040 2041 2042 2043 2044 2045 2046 2047 2048 2049 2050 2051 2052 2053 2054 2055 2056 | |
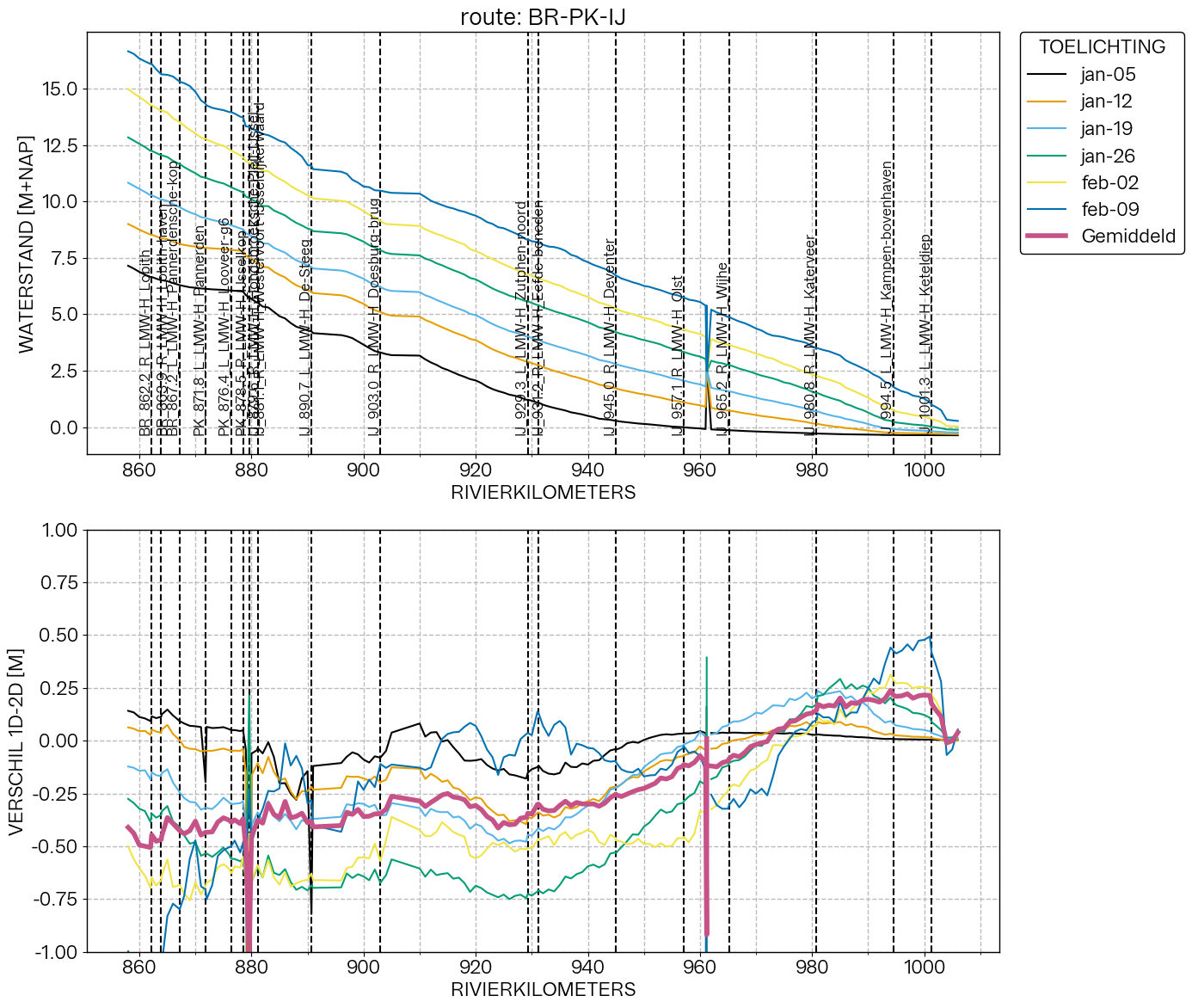
figure_longitudinal(route, stat='time', label='', add_to_fig=None, *, savefig=True)
Create a figure along a route.
Content of figure depends on stat. Figures are saved to [Compare1D2D.output_path]/figures/longitudinal
Example output:
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
route
|
list[str]
|
List of branches (e.g. ['NK', 'LK']) |
required |
stat
|
str
|
What type of longitudinal plot to make (options: "time", "last3", "last25", "max3", |
'time'
|
label
|
str
|
Label of figure. Defaults to "". |
''
|
add_to_fig
|
FigureOutput | None
|
if |
None
|
savefig
|
bool
|
if true, figure is saved to png file. If false, |
True
|
Returns:
| Type | Description |
|---|---|
FigureOutput | None
|
FigureOutput | None: FigureOutput object or None |
Source code in fm2prof\utils.py
2192 2193 2194 2195 2196 2197 2198 2199 2200 2201 2202 2203 2204 2205 2206 2207 2208 2209 2210 2211 2212 2213 2214 2215 2216 2217 2218 2219 2220 2221 2222 2223 2224 2225 2226 2227 2228 2229 2230 2231 2232 2233 2234 2235 2236 2237 2238 2239 2240 2241 2242 2243 2244 2245 2246 2247 2248 2249 2250 2251 2252 2253 2254 2255 2256 2257 2258 2259 2260 2261 2262 2263 2264 2265 2266 2267 2268 2269 2270 2271 2272 2273 2274 2275 2276 2277 2278 2279 2280 2281 2282 2283 2284 2285 2286 2287 2288 2289 2290 2291 2292 2293 2294 2295 2296 | |
figure_longitudinal_rating_curve(route)
Create a figure along a route with lines at various dicharges.
To to this, rating curves are generated at each point by digitizing the model output.
Figures are saved to [Compare1D2D.output_path]/figures/longitudinal
Example output:
.. figure:: figures_utils/longitudinal/example_rating_curve.png
example output figure
Source code in fm2prof\utils.py
2298 2299 2300 2301 2302 2303 2304 2305 2306 2307 2308 2309 2310 2311 2312 2313 2314 2315 2316 2317 2318 2319 2320 2321 2322 2323 2324 2325 2326 2327 2328 2329 2330 2331 2332 2333 2334 2335 2336 2337 2338 2339 2340 2341 2342 2343 2344 2345 2346 2347 2348 2349 2350 2351 2352 2353 2354 2355 2356 2357 2358 2359 2360 2361 2362 2363 2364 2365 2366 2367 2368 2369 2370 2371 | |
figure_longitudinal_time(route)
Create a figure along a route.
Source code in fm2prof\utils.py
get_data_along_route(data, route)
Get data along route.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
data
|
DataFrame
|
DataFrame with data |
required |
route
|
list[str]
|
list with route data |
required |
Returns:
| Type | Description |
|---|---|
DataFrame
|
pd.DataFrame: data |
Source code in fm2prof\utils.py
get_data_along_route_for_time(data, route, time_index)
Get data along route for a given time index.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
data
|
DataFrame
|
Dataframe with data |
required |
route
|
list[str]
|
list of route |
required |
time_index
|
int
|
time index |
required |
Returns:
| Type | Description |
|---|---|
Series
|
pd.Series: Series containing route data |
Source code in fm2prof\utils.py
get_route(route)
Return a sorted list of stations along a route, with rkms.
Source code in fm2prof\utils.py
heatmap_rating_curve(route)
Create a 2D heatmap along a route.
The horizontal axis uses the digitized rating curves to match the two models
Figures are saved to [Compare1D2D.output_path]/figures/heatmap
Example output:
.. figure:: figures_utils/heatmaps/example_rating_curve.png
example output figure
Source code in fm2prof\utils.py
heatmap_time(route)
Create a 2D heatmap along a route.
The horizontal axis uses timemarks to match the 1D and 2D models
Figures are saved to [Compare1D2D.output_path]/figures/heatmap
Example output:
.. figure:: figures_utils/heatmaps/example_time_series.png
example output figure
Source code in fm2prof\utils.py
stations()
statistics_to_file(file_path='error_statistics')
Calculate statistics and write them to file.
The output file is a comma-seperated file with the following columns:
,bias,rkm,branch,is_lmw,std,mae,max13,last25
with for each station:
- bias = bias, mean error
- rkm = river kilometer of the station
- branch = name of 1D branch on which the station lies
- is_lmw = if "LMW" is in the name of station, True.
- std = standard deviation of the rror
- mae = mean absolute error of the error